
Azospirillum sp. RU38E
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Azospirillaceae; Azospirillum; unclassified Azospirillum
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
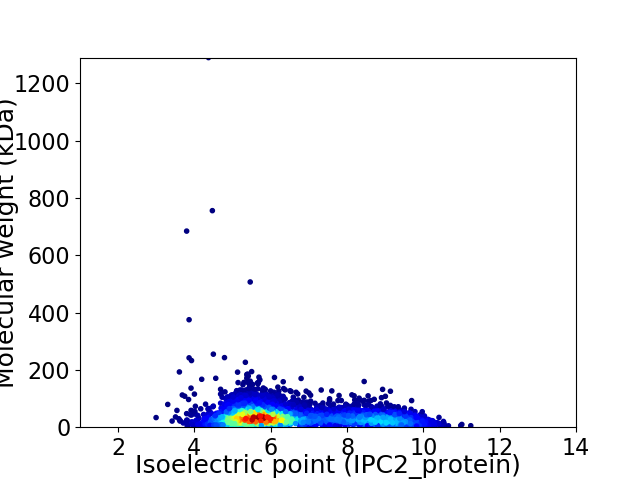
Virtual 2D-PAGE plot for 5137 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A239J751|A0A239J751_9PROT HAMP domain-containing protein OS=Azospirillum sp. RU38E OX=1907313 GN=SAMN05880556_11870 PE=4 SV=1
MM1 pKa = 7.52GIVVTSQSVTGGNGLKK17 pKa = 10.71LSGTATGVVAVSYY30 pKa = 11.37VNGSLLVTEE39 pKa = 5.18GGVAVDD45 pKa = 3.95TYY47 pKa = 11.49GITLPLKK54 pKa = 10.24EE55 pKa = 4.33IDD57 pKa = 3.51ASAVSGMAVSLTADD71 pKa = 3.22VAGGIRR77 pKa = 11.84LIGTGLDD84 pKa = 3.45DD85 pKa = 3.95TLVGGSGNDD94 pKa = 3.62TLIGGAGADD103 pKa = 3.74SLTGGGGADD112 pKa = 4.95DD113 pKa = 5.09FIAYY117 pKa = 10.26NSGTQYY123 pKa = 10.06MPPADD128 pKa = 4.41VITDD132 pKa = 3.82FVVGDD137 pKa = 4.89AIQFGAPVTSIYY149 pKa = 11.02DD150 pKa = 3.46GVRR153 pKa = 11.84RR154 pKa = 11.84SAFDD158 pKa = 3.76GEE160 pKa = 4.33LQVAGNADD168 pKa = 3.3GTTYY172 pKa = 11.05LGLGVSSIVGYY183 pKa = 10.5RR184 pKa = 11.84YY185 pKa = 8.1TLSNSYY191 pKa = 9.91AASAFTIADD200 pKa = 3.64SATGRR205 pKa = 11.84VTLVSQASVTPTEE218 pKa = 4.45GADD221 pKa = 3.6LLTGTTGNDD230 pKa = 3.36SIAGLGGNDD239 pKa = 3.9TIIGLTGSDD248 pKa = 3.87TLDD251 pKa = 3.51GGAGDD256 pKa = 4.96DD257 pKa = 4.35VMRR260 pKa = 11.84QNFGGYY266 pKa = 9.59SILYY270 pKa = 9.35GGAGSDD276 pKa = 3.61TAEE279 pKa = 3.93ITYY282 pKa = 10.72DD283 pKa = 3.55PANGARR289 pKa = 11.84THH291 pKa = 6.83SIVAQFNEE299 pKa = 4.38EE300 pKa = 3.96EE301 pKa = 4.15MSGVRR306 pKa = 11.84INTYY310 pKa = 8.53YY311 pKa = 10.75VSSAVEE317 pKa = 4.06DD318 pKa = 3.79EE319 pKa = 4.73VYY321 pKa = 10.96LIDD324 pKa = 4.46VEE326 pKa = 4.15RR327 pKa = 11.84AIINGVEE334 pKa = 4.02YY335 pKa = 10.44KK336 pKa = 10.35ISNGTLYY343 pKa = 11.29DD344 pKa = 3.68NVLTGAAEE352 pKa = 4.08VRR354 pKa = 11.84NLASGNRR361 pKa = 11.84GADD364 pKa = 3.54SITGGNLQDD373 pKa = 3.57TLYY376 pKa = 10.74GGWGGDD382 pKa = 3.58TLRR385 pKa = 11.84GGAGDD390 pKa = 4.13DD391 pKa = 3.75LLVGGRR397 pKa = 11.84DD398 pKa = 3.32SDD400 pKa = 4.58LLDD403 pKa = 4.1GGAGFDD409 pKa = 4.43RR410 pKa = 11.84AALDD414 pKa = 3.71YY415 pKa = 11.45SEE417 pKa = 4.87NAYY420 pKa = 10.37AVAITVTDD428 pKa = 3.62NNITIDD434 pKa = 4.49NGDD437 pKa = 3.76NDD439 pKa = 3.66QLISIEE445 pKa = 4.13EE446 pKa = 4.13VQVTGTRR453 pKa = 11.84YY454 pKa = 10.57ADD456 pKa = 3.88IITGGNRR463 pKa = 11.84SEE465 pKa = 4.3TLNGGAGADD474 pKa = 4.16LIKK477 pKa = 10.92ACGGADD483 pKa = 3.14ILNGGTGNDD492 pKa = 4.12TLWGGTGDD500 pKa = 4.18DD501 pKa = 4.23LFVLGGRR508 pKa = 11.84RR509 pKa = 11.84SIDD512 pKa = 3.26DD513 pKa = 3.51TDD515 pKa = 3.84YY516 pKa = 11.64VADD519 pKa = 4.51FHH521 pKa = 8.95AGDD524 pKa = 3.69QLSFTHH530 pKa = 6.9RR531 pKa = 11.84LEE533 pKa = 4.09ATEE536 pKa = 5.08LSITSYY542 pKa = 10.72QAGNGSGLQLGQIRR556 pKa = 11.84YY557 pKa = 8.55EE558 pKa = 3.99NLADD562 pKa = 3.7GMVRR566 pKa = 11.84LYY568 pKa = 11.09VGADD572 pKa = 3.4DD573 pKa = 5.9VEE575 pKa = 4.69GADD578 pKa = 3.66YY579 pKa = 10.34TVDD582 pKa = 3.08IAGLGTGALLSISGGNRR599 pKa = 11.84LVLAPTAITNGQGTANADD617 pKa = 3.34IVRR620 pKa = 11.84GAAIDD625 pKa = 3.52TLLIGGKK632 pKa = 10.23GNDD635 pKa = 3.65VIIGGSGTQAAGMEE649 pKa = 4.51ANFADD654 pKa = 4.04VTILRR659 pKa = 11.84DD660 pKa = 3.48ATGVVQVIDD669 pKa = 3.42NRR671 pKa = 11.84ANGEE675 pKa = 4.12GADD678 pKa = 3.66VLSGIEE684 pKa = 4.15LLRR687 pKa = 11.84LNDD690 pKa = 4.66RR691 pKa = 11.84IVLLTDD697 pKa = 3.94PDD699 pKa = 3.87LTSVVPSTTVDD710 pKa = 3.53EE711 pKa = 4.51DD712 pKa = 3.65WYY714 pKa = 10.96LAEE717 pKa = 4.79NPDD720 pKa = 3.54VADD723 pKa = 3.73VVAQGLLQSGAQHH736 pKa = 5.8FQLYY740 pKa = 8.87GQAEE744 pKa = 4.49GRR746 pKa = 11.84QPNAVSDD753 pKa = 3.77NLGNGFSEE761 pKa = 4.18SAYY764 pKa = 10.13LDD766 pKa = 3.46RR767 pKa = 11.84YY768 pKa = 9.93ADD770 pKa = 3.03VRR772 pKa = 11.84AAVNQGIFTSGYY784 pKa = 9.0HH785 pKa = 6.41HH786 pKa = 6.37YY787 pKa = 10.24QVAGRR792 pKa = 11.84AEE794 pKa = 4.02GRR796 pKa = 11.84AGYY799 pKa = 10.16VDD801 pKa = 3.91QGLLAYY807 pKa = 10.07GFDD810 pKa = 3.36EE811 pKa = 4.98SYY813 pKa = 11.58YY814 pKa = 10.8LAANPDD820 pKa = 3.12VAASVRR826 pKa = 11.84VGDD829 pKa = 3.75LASGYY834 pKa = 6.97EE835 pKa = 4.0HH836 pKa = 6.66YY837 pKa = 10.79VRR839 pKa = 11.84SGFAEE844 pKa = 4.18GRR846 pKa = 11.84DD847 pKa = 3.39ANAFFDD853 pKa = 4.11TDD855 pKa = 2.92WYY857 pKa = 10.54LAHH860 pKa = 6.65NPDD863 pKa = 2.62VAAAVQQGVLSSAAMHH879 pKa = 6.57YY880 pKa = 8.07YY881 pKa = 9.51TYY883 pKa = 10.76GWRR886 pKa = 11.84EE887 pKa = 3.57GRR889 pKa = 11.84DD890 pKa = 3.21PSAAFDD896 pKa = 3.68VSAYY900 pKa = 10.47AADD903 pKa = 4.09NPDD906 pKa = 3.01VAAAGINPLLHH917 pKa = 5.76YY918 pKa = 9.96LQYY921 pKa = 10.98GYY923 pKa = 11.24NEE925 pKa = 3.73GRR927 pKa = 11.84IVRR930 pKa = 11.84GLADD934 pKa = 3.81SDD936 pKa = 3.81WAA938 pKa = 4.5
MM1 pKa = 7.52GIVVTSQSVTGGNGLKK17 pKa = 10.71LSGTATGVVAVSYY30 pKa = 11.37VNGSLLVTEE39 pKa = 5.18GGVAVDD45 pKa = 3.95TYY47 pKa = 11.49GITLPLKK54 pKa = 10.24EE55 pKa = 4.33IDD57 pKa = 3.51ASAVSGMAVSLTADD71 pKa = 3.22VAGGIRR77 pKa = 11.84LIGTGLDD84 pKa = 3.45DD85 pKa = 3.95TLVGGSGNDD94 pKa = 3.62TLIGGAGADD103 pKa = 3.74SLTGGGGADD112 pKa = 4.95DD113 pKa = 5.09FIAYY117 pKa = 10.26NSGTQYY123 pKa = 10.06MPPADD128 pKa = 4.41VITDD132 pKa = 3.82FVVGDD137 pKa = 4.89AIQFGAPVTSIYY149 pKa = 11.02DD150 pKa = 3.46GVRR153 pKa = 11.84RR154 pKa = 11.84SAFDD158 pKa = 3.76GEE160 pKa = 4.33LQVAGNADD168 pKa = 3.3GTTYY172 pKa = 11.05LGLGVSSIVGYY183 pKa = 10.5RR184 pKa = 11.84YY185 pKa = 8.1TLSNSYY191 pKa = 9.91AASAFTIADD200 pKa = 3.64SATGRR205 pKa = 11.84VTLVSQASVTPTEE218 pKa = 4.45GADD221 pKa = 3.6LLTGTTGNDD230 pKa = 3.36SIAGLGGNDD239 pKa = 3.9TIIGLTGSDD248 pKa = 3.87TLDD251 pKa = 3.51GGAGDD256 pKa = 4.96DD257 pKa = 4.35VMRR260 pKa = 11.84QNFGGYY266 pKa = 9.59SILYY270 pKa = 9.35GGAGSDD276 pKa = 3.61TAEE279 pKa = 3.93ITYY282 pKa = 10.72DD283 pKa = 3.55PANGARR289 pKa = 11.84THH291 pKa = 6.83SIVAQFNEE299 pKa = 4.38EE300 pKa = 3.96EE301 pKa = 4.15MSGVRR306 pKa = 11.84INTYY310 pKa = 8.53YY311 pKa = 10.75VSSAVEE317 pKa = 4.06DD318 pKa = 3.79EE319 pKa = 4.73VYY321 pKa = 10.96LIDD324 pKa = 4.46VEE326 pKa = 4.15RR327 pKa = 11.84AIINGVEE334 pKa = 4.02YY335 pKa = 10.44KK336 pKa = 10.35ISNGTLYY343 pKa = 11.29DD344 pKa = 3.68NVLTGAAEE352 pKa = 4.08VRR354 pKa = 11.84NLASGNRR361 pKa = 11.84GADD364 pKa = 3.54SITGGNLQDD373 pKa = 3.57TLYY376 pKa = 10.74GGWGGDD382 pKa = 3.58TLRR385 pKa = 11.84GGAGDD390 pKa = 4.13DD391 pKa = 3.75LLVGGRR397 pKa = 11.84DD398 pKa = 3.32SDD400 pKa = 4.58LLDD403 pKa = 4.1GGAGFDD409 pKa = 4.43RR410 pKa = 11.84AALDD414 pKa = 3.71YY415 pKa = 11.45SEE417 pKa = 4.87NAYY420 pKa = 10.37AVAITVTDD428 pKa = 3.62NNITIDD434 pKa = 4.49NGDD437 pKa = 3.76NDD439 pKa = 3.66QLISIEE445 pKa = 4.13EE446 pKa = 4.13VQVTGTRR453 pKa = 11.84YY454 pKa = 10.57ADD456 pKa = 3.88IITGGNRR463 pKa = 11.84SEE465 pKa = 4.3TLNGGAGADD474 pKa = 4.16LIKK477 pKa = 10.92ACGGADD483 pKa = 3.14ILNGGTGNDD492 pKa = 4.12TLWGGTGDD500 pKa = 4.18DD501 pKa = 4.23LFVLGGRR508 pKa = 11.84RR509 pKa = 11.84SIDD512 pKa = 3.26DD513 pKa = 3.51TDD515 pKa = 3.84YY516 pKa = 11.64VADD519 pKa = 4.51FHH521 pKa = 8.95AGDD524 pKa = 3.69QLSFTHH530 pKa = 6.9RR531 pKa = 11.84LEE533 pKa = 4.09ATEE536 pKa = 5.08LSITSYY542 pKa = 10.72QAGNGSGLQLGQIRR556 pKa = 11.84YY557 pKa = 8.55EE558 pKa = 3.99NLADD562 pKa = 3.7GMVRR566 pKa = 11.84LYY568 pKa = 11.09VGADD572 pKa = 3.4DD573 pKa = 5.9VEE575 pKa = 4.69GADD578 pKa = 3.66YY579 pKa = 10.34TVDD582 pKa = 3.08IAGLGTGALLSISGGNRR599 pKa = 11.84LVLAPTAITNGQGTANADD617 pKa = 3.34IVRR620 pKa = 11.84GAAIDD625 pKa = 3.52TLLIGGKK632 pKa = 10.23GNDD635 pKa = 3.65VIIGGSGTQAAGMEE649 pKa = 4.51ANFADD654 pKa = 4.04VTILRR659 pKa = 11.84DD660 pKa = 3.48ATGVVQVIDD669 pKa = 3.42NRR671 pKa = 11.84ANGEE675 pKa = 4.12GADD678 pKa = 3.66VLSGIEE684 pKa = 4.15LLRR687 pKa = 11.84LNDD690 pKa = 4.66RR691 pKa = 11.84IVLLTDD697 pKa = 3.94PDD699 pKa = 3.87LTSVVPSTTVDD710 pKa = 3.53EE711 pKa = 4.51DD712 pKa = 3.65WYY714 pKa = 10.96LAEE717 pKa = 4.79NPDD720 pKa = 3.54VADD723 pKa = 3.73VVAQGLLQSGAQHH736 pKa = 5.8FQLYY740 pKa = 8.87GQAEE744 pKa = 4.49GRR746 pKa = 11.84QPNAVSDD753 pKa = 3.77NLGNGFSEE761 pKa = 4.18SAYY764 pKa = 10.13LDD766 pKa = 3.46RR767 pKa = 11.84YY768 pKa = 9.93ADD770 pKa = 3.03VRR772 pKa = 11.84AAVNQGIFTSGYY784 pKa = 9.0HH785 pKa = 6.41HH786 pKa = 6.37YY787 pKa = 10.24QVAGRR792 pKa = 11.84AEE794 pKa = 4.02GRR796 pKa = 11.84AGYY799 pKa = 10.16VDD801 pKa = 3.91QGLLAYY807 pKa = 10.07GFDD810 pKa = 3.36EE811 pKa = 4.98SYY813 pKa = 11.58YY814 pKa = 10.8LAANPDD820 pKa = 3.12VAASVRR826 pKa = 11.84VGDD829 pKa = 3.75LASGYY834 pKa = 6.97EE835 pKa = 4.0HH836 pKa = 6.66YY837 pKa = 10.79VRR839 pKa = 11.84SGFAEE844 pKa = 4.18GRR846 pKa = 11.84DD847 pKa = 3.39ANAFFDD853 pKa = 4.11TDD855 pKa = 2.92WYY857 pKa = 10.54LAHH860 pKa = 6.65NPDD863 pKa = 2.62VAAAVQQGVLSSAAMHH879 pKa = 6.57YY880 pKa = 8.07YY881 pKa = 9.51TYY883 pKa = 10.76GWRR886 pKa = 11.84EE887 pKa = 3.57GRR889 pKa = 11.84DD890 pKa = 3.21PSAAFDD896 pKa = 3.68VSAYY900 pKa = 10.47AADD903 pKa = 4.09NPDD906 pKa = 3.01VAAAGINPLLHH917 pKa = 5.76YY918 pKa = 9.96LQYY921 pKa = 10.98GYY923 pKa = 11.24NEE925 pKa = 3.73GRR927 pKa = 11.84IVRR930 pKa = 11.84GLADD934 pKa = 3.81SDD936 pKa = 3.81WAA938 pKa = 4.5
Molecular weight: 96.63 kDa
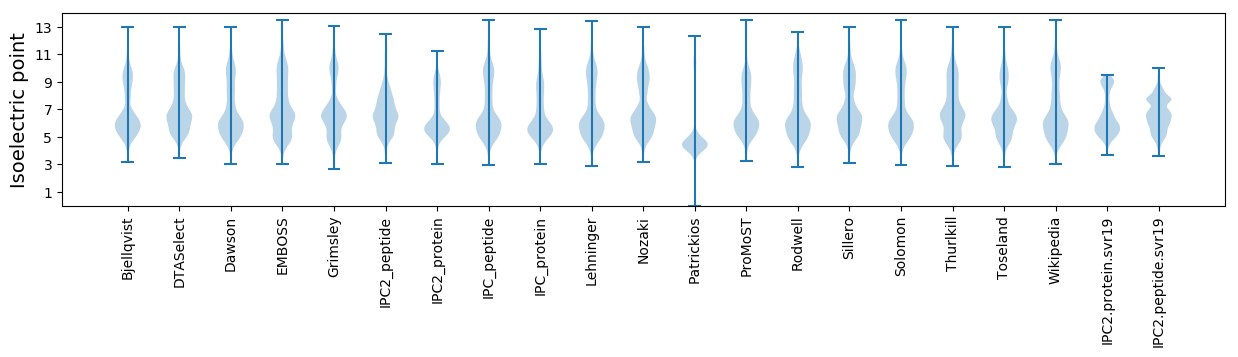
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A239CR87|A0A239CR87_9PROT ATP synthase subunit a OS=Azospirillum sp. RU38E OX=1907313 GN=atpB PE=3 SV=1
MM1 pKa = 7.49KK2 pKa = 10.41RR3 pKa = 11.84PFQPSKK9 pKa = 9.2IVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.49GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVAGRR28 pKa = 11.84NIINKK33 pKa = 9.09RR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.09RR41 pKa = 11.84LSAA44 pKa = 3.93
MM1 pKa = 7.49KK2 pKa = 10.41RR3 pKa = 11.84PFQPSKK9 pKa = 9.2IVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.49GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVAGRR28 pKa = 11.84NIINKK33 pKa = 9.09RR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.09RR41 pKa = 11.84LSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1820581 |
25 |
12860 |
354.4 |
38.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.858 ± 0.048 | 0.763 ± 0.011 |
5.807 ± 0.028 | 5.0 ± 0.036 |
3.382 ± 0.023 | 9.071 ± 0.05 |
1.961 ± 0.021 | 4.626 ± 0.022 |
2.918 ± 0.026 | 10.916 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.324 ± 0.02 | 2.744 ± 0.037 |
5.52 ± 0.045 | 3.58 ± 0.025 |
7.048 ± 0.046 | 5.203 ± 0.051 |
5.525 ± 0.073 | 7.139 ± 0.03 |
1.382 ± 0.016 | 2.232 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
