
Rattail cactus necrosis-associated virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
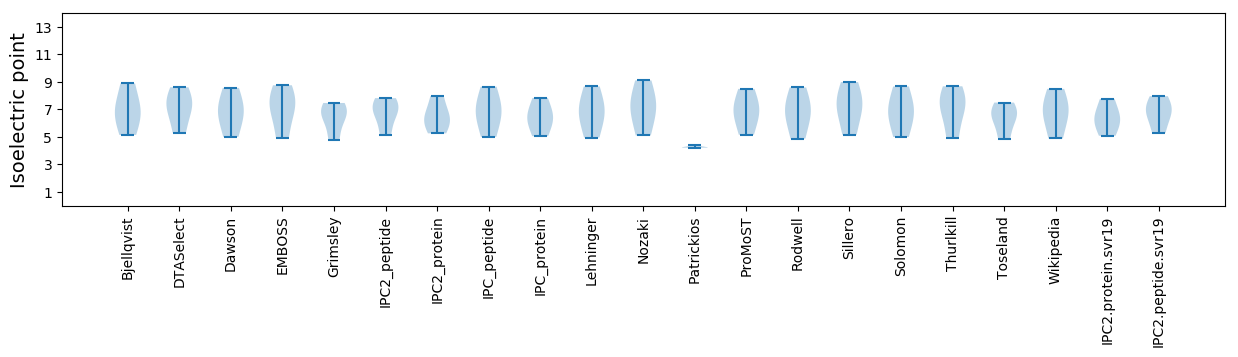
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|G9CU46|G9CU46_9VIRU Capsid protein OS=Rattail cactus necrosis-associated virus OX=1123754 GN=CP PE=3 SV=1
MM1 pKa = 7.94PYY3 pKa = 10.46INVQPKK9 pKa = 10.47DD10 pKa = 3.58FVYY13 pKa = 9.42LTRR16 pKa = 11.84SWVDD20 pKa = 3.28PQRR23 pKa = 11.84LIQFLSDD30 pKa = 3.18MQTQAFQAQQSRR42 pKa = 11.84TQLLNEE48 pKa = 4.59LSTMVVYY55 pKa = 10.75GPTKK59 pKa = 10.32SDD61 pKa = 3.31RR62 pKa = 11.84FPIDD66 pKa = 3.04TYY68 pKa = 11.11LINITKK74 pKa = 10.31GGLSAYY80 pKa = 9.15WFGLTQSADD89 pKa = 3.14TKK91 pKa = 11.25DD92 pKa = 3.25RR93 pKa = 11.84VFEE96 pKa = 4.11VSEE99 pKa = 3.87ARR101 pKa = 11.84AVTNAEE107 pKa = 4.32SKK109 pKa = 10.74LATQRR114 pKa = 11.84VDD116 pKa = 3.29DD117 pKa = 3.91ATVAIRR123 pKa = 11.84NAIKK127 pKa = 9.53STLSYY132 pKa = 10.63LIAGEE137 pKa = 4.72DD138 pKa = 3.28IYY140 pKa = 11.67SRR142 pKa = 11.84TSFEE146 pKa = 5.14AALGWIWQEE155 pKa = 3.94NLPPPPQTAASDD167 pKa = 3.58SRR169 pKa = 11.84TT170 pKa = 3.45
MM1 pKa = 7.94PYY3 pKa = 10.46INVQPKK9 pKa = 10.47DD10 pKa = 3.58FVYY13 pKa = 9.42LTRR16 pKa = 11.84SWVDD20 pKa = 3.28PQRR23 pKa = 11.84LIQFLSDD30 pKa = 3.18MQTQAFQAQQSRR42 pKa = 11.84TQLLNEE48 pKa = 4.59LSTMVVYY55 pKa = 10.75GPTKK59 pKa = 10.32SDD61 pKa = 3.31RR62 pKa = 11.84FPIDD66 pKa = 3.04TYY68 pKa = 11.11LINITKK74 pKa = 10.31GGLSAYY80 pKa = 9.15WFGLTQSADD89 pKa = 3.14TKK91 pKa = 11.25DD92 pKa = 3.25RR93 pKa = 11.84VFEE96 pKa = 4.11VSEE99 pKa = 3.87ARR101 pKa = 11.84AVTNAEE107 pKa = 4.32SKK109 pKa = 10.74LATQRR114 pKa = 11.84VDD116 pKa = 3.29DD117 pKa = 3.91ATVAIRR123 pKa = 11.84NAIKK127 pKa = 9.53STLSYY132 pKa = 10.63LIAGEE137 pKa = 4.72DD138 pKa = 3.28IYY140 pKa = 11.67SRR142 pKa = 11.84TSFEE146 pKa = 5.14AALGWIWQEE155 pKa = 3.94NLPPPPQTAASDD167 pKa = 3.58SRR169 pKa = 11.84TT170 pKa = 3.45
Molecular weight: 19.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G9CU46|G9CU46_9VIRU Capsid protein OS=Rattail cactus necrosis-associated virus OX=1123754 GN=CP PE=3 SV=1
MM1 pKa = 7.82SSLCVSDD8 pKa = 3.95PAVLLSPEE16 pKa = 4.19LHH18 pKa = 6.63LKK20 pKa = 10.05VPQLKK25 pKa = 10.57SLVQFWKK32 pKa = 10.7KK33 pKa = 10.02PFKK36 pKa = 9.84TCSISMSDD44 pKa = 3.84VIKK47 pKa = 10.71VASSNPLSIAVDD59 pKa = 4.6FSSAMPSGPRR69 pKa = 11.84NFKK72 pKa = 10.0YY73 pKa = 10.74VYY75 pKa = 9.66ILAVVLTGRR84 pKa = 11.84WHH86 pKa = 7.06LSEE89 pKa = 4.59SCPGGATFVLYY100 pKa = 10.41DD101 pKa = 3.47KK102 pKa = 10.31RR103 pKa = 11.84LCGRR107 pKa = 11.84AEE109 pKa = 4.4GIYY112 pKa = 10.69GSFFSKK118 pKa = 10.41VSASKK123 pKa = 9.27FQVRR127 pKa = 11.84FSVGHH132 pKa = 5.88SMTVNDD138 pKa = 4.67LLRR141 pKa = 11.84NPLHH145 pKa = 6.66LCFSLEE151 pKa = 4.15SVPCQEE157 pKa = 4.5GYY159 pKa = 10.84EE160 pKa = 4.07PLSVEE165 pKa = 4.25VSSLQLFTDD174 pKa = 5.29CILEE178 pKa = 4.11EE179 pKa = 4.21SLSAKK184 pKa = 9.2LVKK187 pKa = 10.65YY188 pKa = 9.06PALCTDD194 pKa = 3.17NMSVISNVDD203 pKa = 3.21DD204 pKa = 4.61VIFKK208 pKa = 10.53FNSVLNVDD216 pKa = 3.91NVPNSVKK223 pKa = 10.74NVVKK227 pKa = 10.49CNKK230 pKa = 8.47VFKK233 pKa = 10.35GRR235 pKa = 11.84RR236 pKa = 11.84SRR238 pKa = 11.84GKK240 pKa = 8.77WVGSQNEE247 pKa = 4.35VVVKK251 pKa = 10.65DD252 pKa = 3.73GTNVTPHH259 pKa = 6.86RR260 pKa = 11.84LIAKK264 pKa = 8.7DD265 pKa = 3.66ALHH268 pKa = 6.07QRR270 pKa = 11.84TTEE273 pKa = 3.96RR274 pKa = 11.84LRR276 pKa = 11.84LPDD279 pKa = 3.38QVMGRR284 pKa = 11.84PPASDD289 pKa = 3.1TVSEE293 pKa = 4.97RR294 pKa = 11.84YY295 pKa = 9.64ADD297 pKa = 3.59SGFPSTAVSHH307 pKa = 5.25TTSEE311 pKa = 4.11RR312 pKa = 11.84TVDD315 pKa = 3.85DD316 pKa = 4.01GGLRR320 pKa = 11.84SDD322 pKa = 3.99QIGPFPYY329 pKa = 9.72RR330 pKa = 11.84YY331 pKa = 9.8VPYY334 pKa = 10.81
MM1 pKa = 7.82SSLCVSDD8 pKa = 3.95PAVLLSPEE16 pKa = 4.19LHH18 pKa = 6.63LKK20 pKa = 10.05VPQLKK25 pKa = 10.57SLVQFWKK32 pKa = 10.7KK33 pKa = 10.02PFKK36 pKa = 9.84TCSISMSDD44 pKa = 3.84VIKK47 pKa = 10.71VASSNPLSIAVDD59 pKa = 4.6FSSAMPSGPRR69 pKa = 11.84NFKK72 pKa = 10.0YY73 pKa = 10.74VYY75 pKa = 9.66ILAVVLTGRR84 pKa = 11.84WHH86 pKa = 7.06LSEE89 pKa = 4.59SCPGGATFVLYY100 pKa = 10.41DD101 pKa = 3.47KK102 pKa = 10.31RR103 pKa = 11.84LCGRR107 pKa = 11.84AEE109 pKa = 4.4GIYY112 pKa = 10.69GSFFSKK118 pKa = 10.41VSASKK123 pKa = 9.27FQVRR127 pKa = 11.84FSVGHH132 pKa = 5.88SMTVNDD138 pKa = 4.67LLRR141 pKa = 11.84NPLHH145 pKa = 6.66LCFSLEE151 pKa = 4.15SVPCQEE157 pKa = 4.5GYY159 pKa = 10.84EE160 pKa = 4.07PLSVEE165 pKa = 4.25VSSLQLFTDD174 pKa = 5.29CILEE178 pKa = 4.11EE179 pKa = 4.21SLSAKK184 pKa = 9.2LVKK187 pKa = 10.65YY188 pKa = 9.06PALCTDD194 pKa = 3.17NMSVISNVDD203 pKa = 3.21DD204 pKa = 4.61VIFKK208 pKa = 10.53FNSVLNVDD216 pKa = 3.91NVPNSVKK223 pKa = 10.74NVVKK227 pKa = 10.49CNKK230 pKa = 8.47VFKK233 pKa = 10.35GRR235 pKa = 11.84RR236 pKa = 11.84SRR238 pKa = 11.84GKK240 pKa = 8.77WVGSQNEE247 pKa = 4.35VVVKK251 pKa = 10.65DD252 pKa = 3.73GTNVTPHH259 pKa = 6.86RR260 pKa = 11.84LIAKK264 pKa = 8.7DD265 pKa = 3.66ALHH268 pKa = 6.07QRR270 pKa = 11.84TTEE273 pKa = 3.96RR274 pKa = 11.84LRR276 pKa = 11.84LPDD279 pKa = 3.38QVMGRR284 pKa = 11.84PPASDD289 pKa = 3.1TVSEE293 pKa = 4.97RR294 pKa = 11.84YY295 pKa = 9.64ADD297 pKa = 3.59SGFPSTAVSHH307 pKa = 5.25TTSEE311 pKa = 4.11RR312 pKa = 11.84TVDD315 pKa = 3.85DD316 pKa = 4.01GGLRR320 pKa = 11.84SDD322 pKa = 3.99QIGPFPYY329 pKa = 9.72RR330 pKa = 11.84YY331 pKa = 9.8VPYY334 pKa = 10.81
Molecular weight: 36.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3292 |
170 |
1642 |
823.0 |
92.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.652 ± 0.459 | 2.309 ± 0.34 |
6.713 ± 0.38 | 4.465 ± 0.157 |
5.437 ± 0.296 | 5.498 ± 0.357 |
2.552 ± 0.454 | 4.253 ± 0.33 |
5.65 ± 0.319 | 9.022 ± 0.259 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.853 ± 0.072 | 4.587 ± 0.248 |
4.617 ± 0.39 | 3.645 ± 0.578 |
5.437 ± 0.083 | 7.442 ± 1.148 |
5.68 ± 0.542 | 9.721 ± 0.625 |
1.397 ± 0.142 | 3.038 ± 0.176 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
