
Torque teno felis virus
Taxonomy: Viruses; Anelloviridae; Etatorquevirus; Torque teno felid virus 1
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
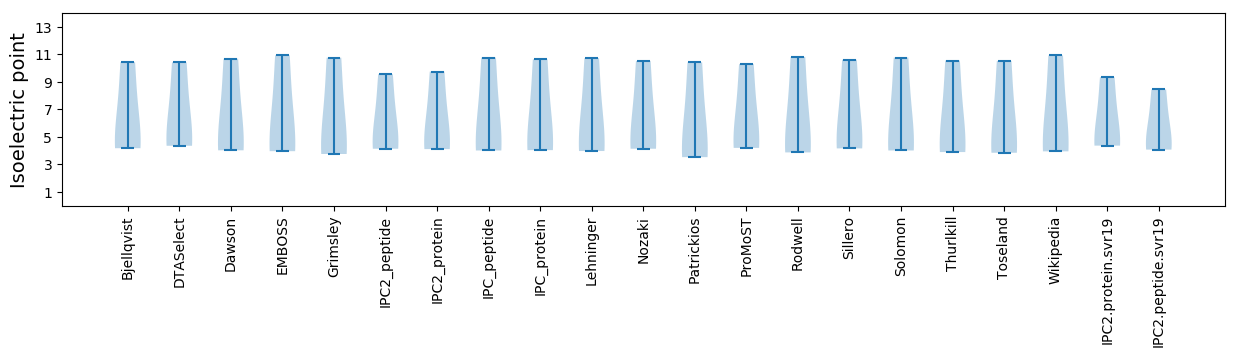
* You can choose from 21 different methods for calculating isoelectric point
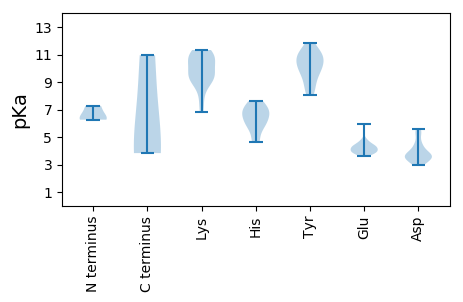
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0D5BV89|A0A0D5BV89_9VIRU Capsid protein OS=Torque teno felis virus OX=687384 PE=3 SV=1
VV1 pKa = 6.26MGSSKK6 pKa = 9.25TRR8 pKa = 11.84LMNEE12 pKa = 3.75LLEE15 pKa = 4.81IIQALSGPDD24 pKa = 3.16PWEE27 pKa = 4.19SDD29 pKa = 2.93GGMGPQGEE37 pKa = 4.25PSKK40 pKa = 11.33SNSRR44 pKa = 11.84LLYY47 pKa = 10.24SDD49 pKa = 5.45PDD51 pKa = 4.19PEE53 pKa = 5.97DD54 pKa = 3.74SSSDD58 pKa = 3.68DD59 pKa = 3.58SEE61 pKa = 3.9MSYY64 pKa = 10.96FSSSDD69 pKa = 3.3STDD72 pKa = 3.59ADD74 pKa = 3.16ITNPNMEE81 pKa = 4.76GGRR84 pKa = 11.84TPPRR88 pKa = 11.84YY89 pKa = 9.09PPRR92 pKa = 11.84TWAFTFKK99 pKa = 10.95
VV1 pKa = 6.26MGSSKK6 pKa = 9.25TRR8 pKa = 11.84LMNEE12 pKa = 3.75LLEE15 pKa = 4.81IIQALSGPDD24 pKa = 3.16PWEE27 pKa = 4.19SDD29 pKa = 2.93GGMGPQGEE37 pKa = 4.25PSKK40 pKa = 11.33SNSRR44 pKa = 11.84LLYY47 pKa = 10.24SDD49 pKa = 5.45PDD51 pKa = 4.19PEE53 pKa = 5.97DD54 pKa = 3.74SSSDD58 pKa = 3.68DD59 pKa = 3.58SEE61 pKa = 3.9MSYY64 pKa = 10.96FSSSDD69 pKa = 3.3STDD72 pKa = 3.59ADD74 pKa = 3.16ITNPNMEE81 pKa = 4.76GGRR84 pKa = 11.84TPPRR88 pKa = 11.84YY89 pKa = 9.09PPRR92 pKa = 11.84TWAFTFKK99 pKa = 10.95
Molecular weight: 10.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D5BV89|A0A0D5BV89_9VIRU Capsid protein OS=Torque teno felis virus OX=687384 PE=3 SV=1
MM1 pKa = 6.38YY2 pKa = 9.92QGRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84WGRR14 pKa = 11.84RR15 pKa = 11.84VLSHH19 pKa = 6.66YY20 pKa = 8.89KK21 pKa = 10.08RR22 pKa = 11.84KK23 pKa = 8.33WRR25 pKa = 11.84RR26 pKa = 11.84GSRR29 pKa = 11.84RR30 pKa = 11.84GHH32 pKa = 6.11GIYY35 pKa = 10.12RR36 pKa = 11.84RR37 pKa = 11.84WRR39 pKa = 11.84RR40 pKa = 11.84WRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84PRR46 pKa = 11.84TVVTEE51 pKa = 3.91QHH53 pKa = 5.75SKK55 pKa = 9.79RR56 pKa = 11.84VKK58 pKa = 9.17TIIVRR63 pKa = 11.84GWEE66 pKa = 3.94PLGNICPTDD75 pKa = 3.53SAKK78 pKa = 10.95AKK80 pKa = 6.83ATPYY84 pKa = 10.93ASYY87 pKa = 11.07DD88 pKa = 3.47SDD90 pKa = 3.65SGQGQWHH97 pKa = 5.87GTWGHH102 pKa = 5.63HH103 pKa = 4.66WFTFQSLVDD112 pKa = 3.63RR113 pKa = 11.84AEE115 pKa = 3.8ARR117 pKa = 11.84LNSFSGNWEE126 pKa = 3.92SYY128 pKa = 10.83DD129 pKa = 3.5YY130 pKa = 11.52LRR132 pKa = 11.84FLGGTMYY139 pKa = 11.33LMQPRR144 pKa = 11.84EE145 pKa = 3.91MCFMFGNDD153 pKa = 4.22PYY155 pKa = 11.86LMTSDD160 pKa = 4.11LDD162 pKa = 3.64KK163 pKa = 10.85TASQKK168 pKa = 10.16NRR170 pKa = 11.84AEE172 pKa = 4.35EE173 pKa = 3.9TWITPGYY180 pKa = 9.47LIHH183 pKa = 7.6RR184 pKa = 11.84PGTHH188 pKa = 7.54LILSRR193 pKa = 11.84QKK195 pKa = 9.31VEE197 pKa = 3.63RR198 pKa = 11.84RR199 pKa = 11.84SMYY202 pKa = 10.04KK203 pKa = 9.58IRR205 pKa = 11.84VPVPTSWRR213 pKa = 11.84GWFPIPEE220 pKa = 4.32CFSYY224 pKa = 10.5VLCHH228 pKa = 7.11WYY230 pKa = 8.44RR231 pKa = 11.84TWWDD235 pKa = 3.18PDD237 pKa = 3.07ACFFDD242 pKa = 4.19PCATGSSCEE251 pKa = 3.99AEE253 pKa = 4.5PWWSTAHH260 pKa = 6.38TKK262 pKa = 8.74QAWVDD267 pKa = 3.57RR268 pKa = 11.84TKK270 pKa = 11.23LDD272 pKa = 3.96DD273 pKa = 4.54PPVGGIGPNQKK284 pKa = 7.54TWAPFLPSRR293 pKa = 11.84PCTNYY298 pKa = 8.33YY299 pKa = 8.02THH301 pKa = 7.08SASFWFKK308 pKa = 11.17YY309 pKa = 9.82KK310 pKa = 10.85LKK312 pKa = 10.6FQVTGEE318 pKa = 4.46NIWAPVPRR326 pKa = 11.84DD327 pKa = 3.29YY328 pKa = 11.19SQQGTVPTAPSRR340 pKa = 11.84QQVEE344 pKa = 4.36SQARR348 pKa = 11.84AAYY351 pKa = 9.9SKK353 pKa = 11.17ANRR356 pKa = 11.84PPTTADD362 pKa = 3.45ILPGDD367 pKa = 4.5LDD369 pKa = 3.8SDD371 pKa = 4.87GILKK375 pKa = 10.64DD376 pKa = 3.4EE377 pKa = 4.56AYY379 pKa = 10.44EE380 pKa = 4.39RR381 pKa = 11.84ITRR384 pKa = 11.84NNPSPKK390 pKa = 9.25RR391 pKa = 11.84PRR393 pKa = 11.84PVGIRR398 pKa = 11.84WRR400 pKa = 11.84DD401 pKa = 3.47GTPGRR406 pKa = 11.84TLQEE410 pKa = 3.87QQQAAVLRR418 pKa = 11.84PRR420 pKa = 11.84PRR422 pKa = 11.84RR423 pKa = 11.84QLLRR427 pKa = 11.84RR428 pKa = 11.84LRR430 pKa = 11.84NVLLQLL436 pKa = 3.99
MM1 pKa = 6.38YY2 pKa = 9.92QGRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84WGRR14 pKa = 11.84RR15 pKa = 11.84VLSHH19 pKa = 6.66YY20 pKa = 8.89KK21 pKa = 10.08RR22 pKa = 11.84KK23 pKa = 8.33WRR25 pKa = 11.84RR26 pKa = 11.84GSRR29 pKa = 11.84RR30 pKa = 11.84GHH32 pKa = 6.11GIYY35 pKa = 10.12RR36 pKa = 11.84RR37 pKa = 11.84WRR39 pKa = 11.84RR40 pKa = 11.84WRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84PRR46 pKa = 11.84TVVTEE51 pKa = 3.91QHH53 pKa = 5.75SKK55 pKa = 9.79RR56 pKa = 11.84VKK58 pKa = 9.17TIIVRR63 pKa = 11.84GWEE66 pKa = 3.94PLGNICPTDD75 pKa = 3.53SAKK78 pKa = 10.95AKK80 pKa = 6.83ATPYY84 pKa = 10.93ASYY87 pKa = 11.07DD88 pKa = 3.47SDD90 pKa = 3.65SGQGQWHH97 pKa = 5.87GTWGHH102 pKa = 5.63HH103 pKa = 4.66WFTFQSLVDD112 pKa = 3.63RR113 pKa = 11.84AEE115 pKa = 3.8ARR117 pKa = 11.84LNSFSGNWEE126 pKa = 3.92SYY128 pKa = 10.83DD129 pKa = 3.5YY130 pKa = 11.52LRR132 pKa = 11.84FLGGTMYY139 pKa = 11.33LMQPRR144 pKa = 11.84EE145 pKa = 3.91MCFMFGNDD153 pKa = 4.22PYY155 pKa = 11.86LMTSDD160 pKa = 4.11LDD162 pKa = 3.64KK163 pKa = 10.85TASQKK168 pKa = 10.16NRR170 pKa = 11.84AEE172 pKa = 4.35EE173 pKa = 3.9TWITPGYY180 pKa = 9.47LIHH183 pKa = 7.6RR184 pKa = 11.84PGTHH188 pKa = 7.54LILSRR193 pKa = 11.84QKK195 pKa = 9.31VEE197 pKa = 3.63RR198 pKa = 11.84RR199 pKa = 11.84SMYY202 pKa = 10.04KK203 pKa = 9.58IRR205 pKa = 11.84VPVPTSWRR213 pKa = 11.84GWFPIPEE220 pKa = 4.32CFSYY224 pKa = 10.5VLCHH228 pKa = 7.11WYY230 pKa = 8.44RR231 pKa = 11.84TWWDD235 pKa = 3.18PDD237 pKa = 3.07ACFFDD242 pKa = 4.19PCATGSSCEE251 pKa = 3.99AEE253 pKa = 4.5PWWSTAHH260 pKa = 6.38TKK262 pKa = 8.74QAWVDD267 pKa = 3.57RR268 pKa = 11.84TKK270 pKa = 11.23LDD272 pKa = 3.96DD273 pKa = 4.54PPVGGIGPNQKK284 pKa = 7.54TWAPFLPSRR293 pKa = 11.84PCTNYY298 pKa = 8.33YY299 pKa = 8.02THH301 pKa = 7.08SASFWFKK308 pKa = 11.17YY309 pKa = 9.82KK310 pKa = 10.85LKK312 pKa = 10.6FQVTGEE318 pKa = 4.46NIWAPVPRR326 pKa = 11.84DD327 pKa = 3.29YY328 pKa = 11.19SQQGTVPTAPSRR340 pKa = 11.84QQVEE344 pKa = 4.36SQARR348 pKa = 11.84AAYY351 pKa = 9.9SKK353 pKa = 11.17ANRR356 pKa = 11.84PPTTADD362 pKa = 3.45ILPGDD367 pKa = 4.5LDD369 pKa = 3.8SDD371 pKa = 4.87GILKK375 pKa = 10.64DD376 pKa = 3.4EE377 pKa = 4.56AYY379 pKa = 10.44EE380 pKa = 4.39RR381 pKa = 11.84ITRR384 pKa = 11.84NNPSPKK390 pKa = 9.25RR391 pKa = 11.84PRR393 pKa = 11.84PVGIRR398 pKa = 11.84WRR400 pKa = 11.84DD401 pKa = 3.47GTPGRR406 pKa = 11.84TLQEE410 pKa = 3.87QQQAAVLRR418 pKa = 11.84PRR420 pKa = 11.84PRR422 pKa = 11.84RR423 pKa = 11.84QLLRR427 pKa = 11.84RR428 pKa = 11.84LRR430 pKa = 11.84NVLLQLL436 pKa = 3.99
Molecular weight: 51.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
640 |
99 |
436 |
213.3 |
24.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.156 ± 0.678 | 1.719 ± 0.487 |
6.563 ± 1.789 | 5.313 ± 1.727 |
3.281 ± 0.139 | 7.187 ± 0.549 |
2.5 ± 0.772 | 3.281 ± 0.163 |
4.219 ± 0.325 | 6.25 ± 0.11 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.188 ± 0.819 | 2.656 ± 0.5 |
7.813 ± 1.181 | 4.375 ± 0.771 |
10.156 ± 2.75 | 8.906 ± 2.642 |
6.094 ± 0.92 | 4.375 ± 1.066 |
4.063 ± 1.008 | 3.906 ± 0.462 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
