
Micrococcales bacterium KH10
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; unclassified Micrococcales
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
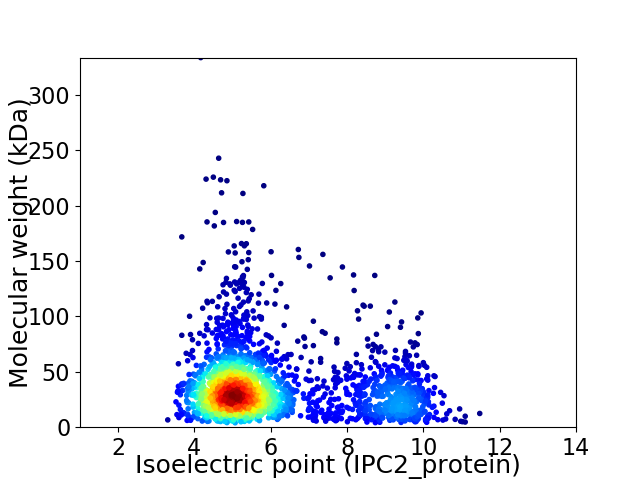
Virtual 2D-PAGE plot for 2348 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A239GDC6|A0A239GDC6_9MICO Histidine kinase OS=Micrococcales bacterium KH10 OX=1945885 GN=SAMN06309944_1268 PE=4 SV=1
MM1 pKa = 7.32SRR3 pKa = 11.84IKK5 pKa = 11.05NPFSVNRR12 pKa = 11.84VATASIATGLALVLTACGGGTDD34 pKa = 3.93SPTPNDD40 pKa = 2.96VATNTEE46 pKa = 4.32GQSEE50 pKa = 4.35EE51 pKa = 4.08LSGEE55 pKa = 4.27VTLIAHH61 pKa = 7.43DD62 pKa = 4.14SFTFPDD68 pKa = 3.54EE69 pKa = 4.8LIADD73 pKa = 4.49FEE75 pKa = 4.8EE76 pKa = 4.76KK77 pKa = 9.97TGLTLNVTTTGDD89 pKa = 3.37GGQLANQLVLTKK101 pKa = 10.38DD102 pKa = 3.29APLADD107 pKa = 3.58AFFGVDD113 pKa = 3.08NSFVTGLIEE122 pKa = 4.77AGAVDD127 pKa = 5.13PYY129 pKa = 10.62TPEE132 pKa = 3.98SLGEE136 pKa = 4.15SATEE140 pKa = 4.24FAADD144 pKa = 3.92DD145 pKa = 4.51AGSVTPVDD153 pKa = 3.11HH154 pKa = 7.59GYY156 pKa = 10.62VCFNADD162 pKa = 3.43DD163 pKa = 3.56AWFEE167 pKa = 4.46EE168 pKa = 4.4NGLAAPEE175 pKa = 4.16TLDD178 pKa = 4.83DD179 pKa = 4.51LTDD182 pKa = 3.25EE183 pKa = 4.97AYY185 pKa = 10.44RR186 pKa = 11.84GLTVVMDD193 pKa = 4.49PTASSPGAAMLFATVAKK210 pKa = 10.13FGEE213 pKa = 4.58DD214 pKa = 3.37GYY216 pKa = 11.37RR217 pKa = 11.84DD218 pKa = 3.74YY219 pKa = 9.1WTKK222 pKa = 11.21LLANDD227 pKa = 3.93TKK229 pKa = 10.99VVQGWSDD236 pKa = 3.06AYY238 pKa = 11.21YY239 pKa = 10.95VDD241 pKa = 4.67FSGGGEE247 pKa = 4.03DD248 pKa = 3.39GQYY251 pKa = 10.45PIVVSYY257 pKa = 8.33NTSPAYY263 pKa = 9.13TVAEE267 pKa = 4.56GSDD270 pKa = 3.35EE271 pKa = 4.32STTSVLLDD279 pKa = 3.17TCTRR283 pKa = 11.84QVEE286 pKa = 4.48YY287 pKa = 11.1AGVLAGAKK295 pKa = 9.78NPAGARR301 pKa = 11.84AVIDD305 pKa = 3.68YY306 pKa = 10.1LVSQDD311 pKa = 3.83FQDD314 pKa = 4.01TVAEE318 pKa = 4.13TMYY321 pKa = 9.27MYY323 pKa = 10.57PVLPSGAVPDD333 pKa = 4.43EE334 pKa = 4.18WQNWAPVPQHH344 pKa = 6.64SLDD347 pKa = 3.86LAPDD351 pKa = 3.56QITANRR357 pKa = 11.84AAWLDD362 pKa = 3.44EE363 pKa = 3.79WSQLAGG369 pKa = 3.45
MM1 pKa = 7.32SRR3 pKa = 11.84IKK5 pKa = 11.05NPFSVNRR12 pKa = 11.84VATASIATGLALVLTACGGGTDD34 pKa = 3.93SPTPNDD40 pKa = 2.96VATNTEE46 pKa = 4.32GQSEE50 pKa = 4.35EE51 pKa = 4.08LSGEE55 pKa = 4.27VTLIAHH61 pKa = 7.43DD62 pKa = 4.14SFTFPDD68 pKa = 3.54EE69 pKa = 4.8LIADD73 pKa = 4.49FEE75 pKa = 4.8EE76 pKa = 4.76KK77 pKa = 9.97TGLTLNVTTTGDD89 pKa = 3.37GGQLANQLVLTKK101 pKa = 10.38DD102 pKa = 3.29APLADD107 pKa = 3.58AFFGVDD113 pKa = 3.08NSFVTGLIEE122 pKa = 4.77AGAVDD127 pKa = 5.13PYY129 pKa = 10.62TPEE132 pKa = 3.98SLGEE136 pKa = 4.15SATEE140 pKa = 4.24FAADD144 pKa = 3.92DD145 pKa = 4.51AGSVTPVDD153 pKa = 3.11HH154 pKa = 7.59GYY156 pKa = 10.62VCFNADD162 pKa = 3.43DD163 pKa = 3.56AWFEE167 pKa = 4.46EE168 pKa = 4.4NGLAAPEE175 pKa = 4.16TLDD178 pKa = 4.83DD179 pKa = 4.51LTDD182 pKa = 3.25EE183 pKa = 4.97AYY185 pKa = 10.44RR186 pKa = 11.84GLTVVMDD193 pKa = 4.49PTASSPGAAMLFATVAKK210 pKa = 10.13FGEE213 pKa = 4.58DD214 pKa = 3.37GYY216 pKa = 11.37RR217 pKa = 11.84DD218 pKa = 3.74YY219 pKa = 9.1WTKK222 pKa = 11.21LLANDD227 pKa = 3.93TKK229 pKa = 10.99VVQGWSDD236 pKa = 3.06AYY238 pKa = 11.21YY239 pKa = 10.95VDD241 pKa = 4.67FSGGGEE247 pKa = 4.03DD248 pKa = 3.39GQYY251 pKa = 10.45PIVVSYY257 pKa = 8.33NTSPAYY263 pKa = 9.13TVAEE267 pKa = 4.56GSDD270 pKa = 3.35EE271 pKa = 4.32STTSVLLDD279 pKa = 3.17TCTRR283 pKa = 11.84QVEE286 pKa = 4.48YY287 pKa = 11.1AGVLAGAKK295 pKa = 9.78NPAGARR301 pKa = 11.84AVIDD305 pKa = 3.68YY306 pKa = 10.1LVSQDD311 pKa = 3.83FQDD314 pKa = 4.01TVAEE318 pKa = 4.13TMYY321 pKa = 9.27MYY323 pKa = 10.57PVLPSGAVPDD333 pKa = 4.43EE334 pKa = 4.18WQNWAPVPQHH344 pKa = 6.64SLDD347 pKa = 3.86LAPDD351 pKa = 3.56QITANRR357 pKa = 11.84AAWLDD362 pKa = 3.44EE363 pKa = 3.79WSQLAGG369 pKa = 3.45
Molecular weight: 39.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A239E1X9|A0A239E1X9_9MICO Carbamoyl-phosphate synthase small chain OS=Micrococcales bacterium KH10 OX=1945885 GN=carA PE=3 SV=1
AA1 pKa = 7.79RR2 pKa = 11.84GLRR5 pKa = 11.84VVKK8 pKa = 10.46ARR10 pKa = 11.84PKK12 pKa = 10.23KK13 pKa = 8.36VTVRR17 pKa = 11.84TKK19 pKa = 8.73QVRR22 pKa = 11.84SGASVKK28 pKa = 10.24VRR30 pKa = 11.84VKK32 pKa = 10.25VAKK35 pKa = 10.36LNNGRR40 pKa = 11.84HH41 pKa = 5.14ANGRR45 pKa = 11.84VQIRR49 pKa = 11.84IGKK52 pKa = 8.94RR53 pKa = 11.84VIGSVRR59 pKa = 11.84IKK61 pKa = 10.57PKK63 pKa = 10.44AKK65 pKa = 9.89GNRR68 pKa = 11.84TVTVKK73 pKa = 10.67LSSAQRR79 pKa = 11.84SGSSLKK85 pKa = 10.27VRR87 pKa = 11.84AKK89 pKa = 10.32FVAKK93 pKa = 10.38NNKK96 pKa = 8.4SFKK99 pKa = 9.8NRR101 pKa = 11.84QSKK104 pKa = 7.91VTTIRR109 pKa = 11.84LVRR112 pKa = 3.88
AA1 pKa = 7.79RR2 pKa = 11.84GLRR5 pKa = 11.84VVKK8 pKa = 10.46ARR10 pKa = 11.84PKK12 pKa = 10.23KK13 pKa = 8.36VTVRR17 pKa = 11.84TKK19 pKa = 8.73QVRR22 pKa = 11.84SGASVKK28 pKa = 10.24VRR30 pKa = 11.84VKK32 pKa = 10.25VAKK35 pKa = 10.36LNNGRR40 pKa = 11.84HH41 pKa = 5.14ANGRR45 pKa = 11.84VQIRR49 pKa = 11.84IGKK52 pKa = 8.94RR53 pKa = 11.84VIGSVRR59 pKa = 11.84IKK61 pKa = 10.57PKK63 pKa = 10.44AKK65 pKa = 9.89GNRR68 pKa = 11.84TVTVKK73 pKa = 10.67LSSAQRR79 pKa = 11.84SGSSLKK85 pKa = 10.27VRR87 pKa = 11.84AKK89 pKa = 10.32FVAKK93 pKa = 10.38NNKK96 pKa = 8.4SFKK99 pKa = 9.8NRR101 pKa = 11.84QSKK104 pKa = 7.91VTTIRR109 pKa = 11.84LVRR112 pKa = 3.88
Molecular weight: 12.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
839289 |
37 |
3203 |
357.4 |
38.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.13 ± 0.06 | 0.565 ± 0.012 |
6.288 ± 0.041 | 5.515 ± 0.052 |
2.831 ± 0.025 | 8.373 ± 0.043 |
2.033 ± 0.022 | 4.832 ± 0.034 |
2.827 ± 0.053 | 9.246 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.809 ± 0.018 | 2.691 ± 0.041 |
5.15 ± 0.038 | 3.551 ± 0.026 |
6.736 ± 0.053 | 6.205 ± 0.042 |
6.882 ± 0.064 | 8.711 ± 0.042 |
1.47 ± 0.022 | 2.156 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
