
Microbacterium agarici
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
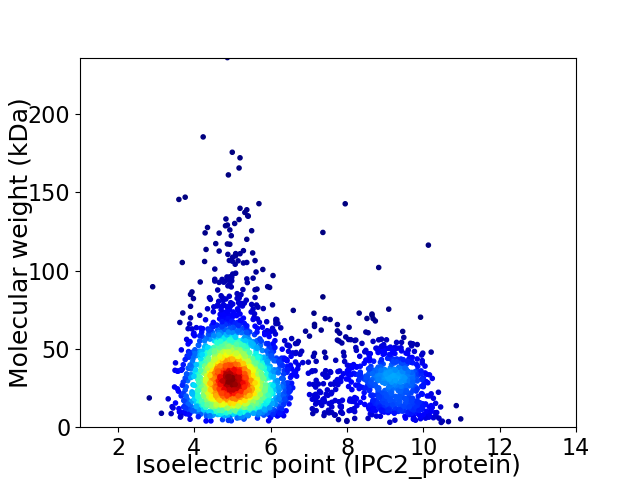
Virtual 2D-PAGE plot for 3074 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A9DVY7|A0A2A9DVY7_9MICO Carbohydrate ABC transporter substrate-binding protein (CUT1 family) OS=Microbacterium agarici OX=630514 GN=ATJ78_1459 PE=4 SV=1
MM1 pKa = 7.11PAARR5 pKa = 11.84RR6 pKa = 11.84VALVSTLAVLGTALIAPTAANAATPPGGVEE36 pKa = 3.99LTLLATTDD44 pKa = 3.63THH46 pKa = 6.24GHH48 pKa = 4.99VADD51 pKa = 3.51WDD53 pKa = 4.11YY54 pKa = 11.19FQNAPYY60 pKa = 8.67PTDD63 pKa = 3.98DD64 pKa = 3.43SLGLSRR70 pKa = 11.84VASVVDD76 pKa = 3.73DD77 pKa = 3.75VRR79 pKa = 11.84AEE81 pKa = 4.02KK82 pKa = 10.67GDD84 pKa = 3.62EE85 pKa = 4.25SVVVLDD91 pKa = 5.03NGDD94 pKa = 4.63AIQGTPLTYY103 pKa = 10.32LYY105 pKa = 10.78GYY107 pKa = 9.96GDD109 pKa = 3.65SRR111 pKa = 11.84DD112 pKa = 3.58SVLNDD117 pKa = 3.9TIEE120 pKa = 4.61HH121 pKa = 6.48PMAQVFDD128 pKa = 4.5TIGYY132 pKa = 7.71DD133 pKa = 3.31AQVVGNHH140 pKa = 5.97EE141 pKa = 4.32YY142 pKa = 11.02NYY144 pKa = 11.25GLDD147 pKa = 3.66LLQAYY152 pKa = 10.09DD153 pKa = 4.3EE154 pKa = 4.84DD155 pKa = 5.43LEE157 pKa = 5.7APLLGANVIDD167 pKa = 4.33AATGEE172 pKa = 4.8PYY174 pKa = 10.24HH175 pKa = 7.3DD176 pKa = 5.51PYY178 pKa = 11.41TLIDD182 pKa = 3.68RR183 pKa = 11.84EE184 pKa = 4.01IDD186 pKa = 3.49GKK188 pKa = 10.46QVTIGVIGLVTPGVRR203 pKa = 11.84IWDD206 pKa = 3.76KK207 pKa = 10.65QHH209 pKa = 6.82VEE211 pKa = 4.72GILEE215 pKa = 4.09FQDD218 pKa = 3.18MVEE221 pKa = 4.6AAKK224 pKa = 10.34QWVPEE229 pKa = 3.83VDD231 pKa = 3.24AQADD235 pKa = 4.03VVVVLAHH242 pKa = 6.55TGQGTVPDD250 pKa = 4.57DD251 pKa = 4.6AYY253 pKa = 11.22DD254 pKa = 3.98PADD257 pKa = 4.02LNEE260 pKa = 4.56DD261 pKa = 3.55VANNIAFQVPGIDD274 pKa = 3.46VLVAGHH280 pKa = 5.82SHH282 pKa = 5.7QDD284 pKa = 3.25EE285 pKa = 4.6PQTLITNTAGEE296 pKa = 4.37QTLITQPNYY305 pKa = 7.76WARR308 pKa = 11.84SVSEE312 pKa = 4.34VTLSLAPTADD322 pKa = 3.21GFEE325 pKa = 4.43VDD327 pKa = 3.88WSDD330 pKa = 3.47GYY332 pKa = 11.81APVVTPHH339 pKa = 6.59YY340 pKa = 9.11GTDD343 pKa = 3.68GTPEE347 pKa = 4.29DD348 pKa = 4.05PQVLEE353 pKa = 4.77VISEE357 pKa = 4.03EE358 pKa = 4.1HH359 pKa = 6.45EE360 pKa = 3.99ATIEE364 pKa = 4.08YY365 pKa = 10.73VNTPVADD372 pKa = 3.85SVEE375 pKa = 4.24EE376 pKa = 4.12LPAATSRR383 pKa = 11.84YY384 pKa = 9.07EE385 pKa = 3.75DD386 pKa = 3.32TAIIDD391 pKa = 4.57FINSIQLEE399 pKa = 4.56TVDD402 pKa = 3.89AALDD406 pKa = 3.61GTEE409 pKa = 4.13YY410 pKa = 11.41ADD412 pKa = 4.01VPVISQASPFSRR424 pKa = 11.84TALFPEE430 pKa = 4.85GEE432 pKa = 4.2VTIRR436 pKa = 11.84DD437 pKa = 3.33IAGLYY442 pKa = 9.73IYY444 pKa = 10.15EE445 pKa = 4.12NTLVGVQMTGAEE457 pKa = 3.62IAEE460 pKa = 4.09YY461 pKa = 10.73LEE463 pKa = 3.87YY464 pKa = 10.19SARR467 pKa = 11.84YY468 pKa = 8.18FVQTEE473 pKa = 4.13PGAEE477 pKa = 3.83FDD479 pKa = 4.32PATGTNAVYY488 pKa = 10.01PDD490 pKa = 4.36RR491 pKa = 11.84PDD493 pKa = 3.43GVPDD497 pKa = 3.47YY498 pKa = 10.99NYY500 pKa = 11.18DD501 pKa = 3.51VLTGLSYY508 pKa = 10.7TIDD511 pKa = 3.01ISQPVGEE518 pKa = 5.25RR519 pKa = 11.84ILDD522 pKa = 3.82LAHH525 pKa = 7.63PDD527 pKa = 3.82GTPVAADD534 pKa = 3.52DD535 pKa = 4.21TFVLAVNNYY544 pKa = 7.8RR545 pKa = 11.84QSGGGAFPNVADD557 pKa = 4.12APVVYY562 pKa = 10.08DD563 pKa = 3.46EE564 pKa = 4.66RR565 pKa = 11.84LEE567 pKa = 3.89IRR569 pKa = 11.84QLLIDD574 pKa = 3.72WASEE578 pKa = 3.76RR579 pKa = 11.84GTIDD583 pKa = 3.18PADD586 pKa = 3.57FFVEE590 pKa = 3.95NWEE593 pKa = 4.28LTTDD597 pKa = 3.75AATEE601 pKa = 4.45TPTPTPEE608 pKa = 3.92PTDD611 pKa = 4.03PGEE614 pKa = 4.49GDD616 pKa = 4.1DD617 pKa = 5.4PGAGDD622 pKa = 5.43DD623 pKa = 4.72GSTDD627 pKa = 3.49APGTGSDD634 pKa = 3.71GAANDD639 pKa = 4.81DD640 pKa = 4.01SSTDD644 pKa = 3.46AASDD648 pKa = 4.0LPVTGADD655 pKa = 3.38IAWPLGLAAALLLLGAGLVATRR677 pKa = 11.84RR678 pKa = 11.84FARR681 pKa = 11.84KK682 pKa = 9.38RR683 pKa = 11.84SS684 pKa = 3.44
MM1 pKa = 7.11PAARR5 pKa = 11.84RR6 pKa = 11.84VALVSTLAVLGTALIAPTAANAATPPGGVEE36 pKa = 3.99LTLLATTDD44 pKa = 3.63THH46 pKa = 6.24GHH48 pKa = 4.99VADD51 pKa = 3.51WDD53 pKa = 4.11YY54 pKa = 11.19FQNAPYY60 pKa = 8.67PTDD63 pKa = 3.98DD64 pKa = 3.43SLGLSRR70 pKa = 11.84VASVVDD76 pKa = 3.73DD77 pKa = 3.75VRR79 pKa = 11.84AEE81 pKa = 4.02KK82 pKa = 10.67GDD84 pKa = 3.62EE85 pKa = 4.25SVVVLDD91 pKa = 5.03NGDD94 pKa = 4.63AIQGTPLTYY103 pKa = 10.32LYY105 pKa = 10.78GYY107 pKa = 9.96GDD109 pKa = 3.65SRR111 pKa = 11.84DD112 pKa = 3.58SVLNDD117 pKa = 3.9TIEE120 pKa = 4.61HH121 pKa = 6.48PMAQVFDD128 pKa = 4.5TIGYY132 pKa = 7.71DD133 pKa = 3.31AQVVGNHH140 pKa = 5.97EE141 pKa = 4.32YY142 pKa = 11.02NYY144 pKa = 11.25GLDD147 pKa = 3.66LLQAYY152 pKa = 10.09DD153 pKa = 4.3EE154 pKa = 4.84DD155 pKa = 5.43LEE157 pKa = 5.7APLLGANVIDD167 pKa = 4.33AATGEE172 pKa = 4.8PYY174 pKa = 10.24HH175 pKa = 7.3DD176 pKa = 5.51PYY178 pKa = 11.41TLIDD182 pKa = 3.68RR183 pKa = 11.84EE184 pKa = 4.01IDD186 pKa = 3.49GKK188 pKa = 10.46QVTIGVIGLVTPGVRR203 pKa = 11.84IWDD206 pKa = 3.76KK207 pKa = 10.65QHH209 pKa = 6.82VEE211 pKa = 4.72GILEE215 pKa = 4.09FQDD218 pKa = 3.18MVEE221 pKa = 4.6AAKK224 pKa = 10.34QWVPEE229 pKa = 3.83VDD231 pKa = 3.24AQADD235 pKa = 4.03VVVVLAHH242 pKa = 6.55TGQGTVPDD250 pKa = 4.57DD251 pKa = 4.6AYY253 pKa = 11.22DD254 pKa = 3.98PADD257 pKa = 4.02LNEE260 pKa = 4.56DD261 pKa = 3.55VANNIAFQVPGIDD274 pKa = 3.46VLVAGHH280 pKa = 5.82SHH282 pKa = 5.7QDD284 pKa = 3.25EE285 pKa = 4.6PQTLITNTAGEE296 pKa = 4.37QTLITQPNYY305 pKa = 7.76WARR308 pKa = 11.84SVSEE312 pKa = 4.34VTLSLAPTADD322 pKa = 3.21GFEE325 pKa = 4.43VDD327 pKa = 3.88WSDD330 pKa = 3.47GYY332 pKa = 11.81APVVTPHH339 pKa = 6.59YY340 pKa = 9.11GTDD343 pKa = 3.68GTPEE347 pKa = 4.29DD348 pKa = 4.05PQVLEE353 pKa = 4.77VISEE357 pKa = 4.03EE358 pKa = 4.1HH359 pKa = 6.45EE360 pKa = 3.99ATIEE364 pKa = 4.08YY365 pKa = 10.73VNTPVADD372 pKa = 3.85SVEE375 pKa = 4.24EE376 pKa = 4.12LPAATSRR383 pKa = 11.84YY384 pKa = 9.07EE385 pKa = 3.75DD386 pKa = 3.32TAIIDD391 pKa = 4.57FINSIQLEE399 pKa = 4.56TVDD402 pKa = 3.89AALDD406 pKa = 3.61GTEE409 pKa = 4.13YY410 pKa = 11.41ADD412 pKa = 4.01VPVISQASPFSRR424 pKa = 11.84TALFPEE430 pKa = 4.85GEE432 pKa = 4.2VTIRR436 pKa = 11.84DD437 pKa = 3.33IAGLYY442 pKa = 9.73IYY444 pKa = 10.15EE445 pKa = 4.12NTLVGVQMTGAEE457 pKa = 3.62IAEE460 pKa = 4.09YY461 pKa = 10.73LEE463 pKa = 3.87YY464 pKa = 10.19SARR467 pKa = 11.84YY468 pKa = 8.18FVQTEE473 pKa = 4.13PGAEE477 pKa = 3.83FDD479 pKa = 4.32PATGTNAVYY488 pKa = 10.01PDD490 pKa = 4.36RR491 pKa = 11.84PDD493 pKa = 3.43GVPDD497 pKa = 3.47YY498 pKa = 10.99NYY500 pKa = 11.18DD501 pKa = 3.51VLTGLSYY508 pKa = 10.7TIDD511 pKa = 3.01ISQPVGEE518 pKa = 5.25RR519 pKa = 11.84ILDD522 pKa = 3.82LAHH525 pKa = 7.63PDD527 pKa = 3.82GTPVAADD534 pKa = 3.52DD535 pKa = 4.21TFVLAVNNYY544 pKa = 7.8RR545 pKa = 11.84QSGGGAFPNVADD557 pKa = 4.12APVVYY562 pKa = 10.08DD563 pKa = 3.46EE564 pKa = 4.66RR565 pKa = 11.84LEE567 pKa = 3.89IRR569 pKa = 11.84QLLIDD574 pKa = 3.72WASEE578 pKa = 3.76RR579 pKa = 11.84GTIDD583 pKa = 3.18PADD586 pKa = 3.57FFVEE590 pKa = 3.95NWEE593 pKa = 4.28LTTDD597 pKa = 3.75AATEE601 pKa = 4.45TPTPTPEE608 pKa = 3.92PTDD611 pKa = 4.03PGEE614 pKa = 4.49GDD616 pKa = 4.1DD617 pKa = 5.4PGAGDD622 pKa = 5.43DD623 pKa = 4.72GSTDD627 pKa = 3.49APGTGSDD634 pKa = 3.71GAANDD639 pKa = 4.81DD640 pKa = 4.01SSTDD644 pKa = 3.46AASDD648 pKa = 4.0LPVTGADD655 pKa = 3.38IAWPLGLAAALLLLGAGLVATRR677 pKa = 11.84RR678 pKa = 11.84FARR681 pKa = 11.84KK682 pKa = 9.38RR683 pKa = 11.84SS684 pKa = 3.44
Molecular weight: 72.93 kDa
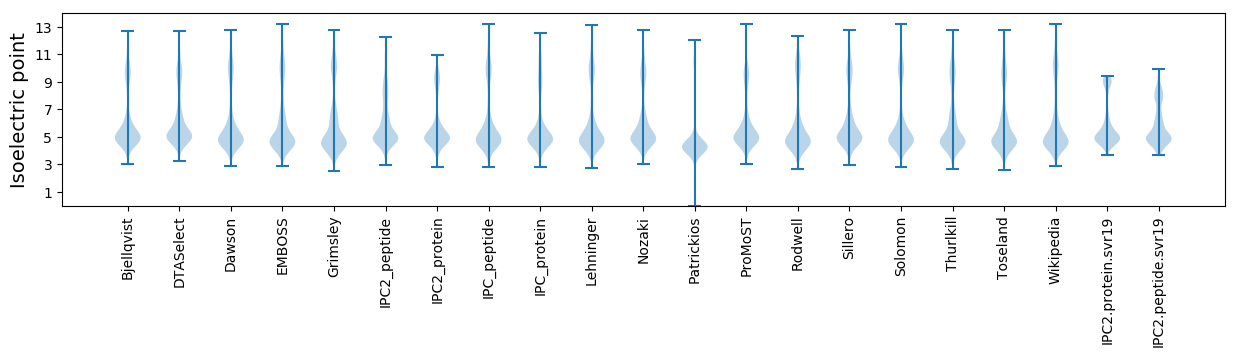
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A9DTK4|A0A2A9DTK4_9MICO Uncharacterized protein OS=Microbacterium agarici OX=630514 GN=ATJ78_0381 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.5VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVSARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.97GRR40 pKa = 11.84TEE42 pKa = 3.99LSAA45 pKa = 4.86
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.5VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVSARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.97GRR40 pKa = 11.84TEE42 pKa = 3.99LSAA45 pKa = 4.86
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
989917 |
29 |
2145 |
322.0 |
34.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.632 ± 0.059 | 0.548 ± 0.011 |
6.48 ± 0.043 | 5.84 ± 0.045 |
3.182 ± 0.026 | 8.484 ± 0.042 |
2.059 ± 0.021 | 4.969 ± 0.034 |
2.237 ± 0.032 | 9.75 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.989 ± 0.019 | 2.256 ± 0.021 |
4.996 ± 0.027 | 2.868 ± 0.021 |
6.998 ± 0.05 | 6.288 ± 0.032 |
6.145 ± 0.033 | 8.832 ± 0.039 |
1.422 ± 0.018 | 2.024 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
