
Puerto Almendras virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Almendravirus; Puerto Almendras almendravirus
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
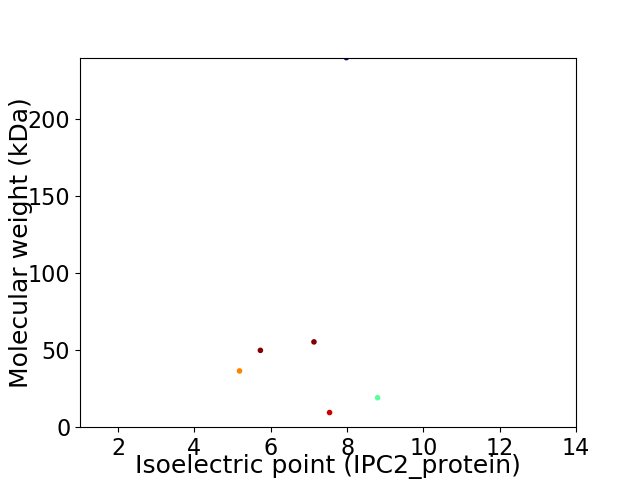
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|X4R562|X4R562_9RHAB P protein OS=Puerto Almendras virus OX=1479613 GN=P PE=4 SV=1
MM1 pKa = 7.43SRR3 pKa = 11.84NHH5 pKa = 5.87FQKK8 pKa = 10.77LGNKK12 pKa = 9.25PDD14 pKa = 3.59IQADD18 pKa = 3.54IYY20 pKa = 11.62DD21 pKa = 4.12SANLMSQINLNAYY34 pKa = 7.05TISKK38 pKa = 9.95RR39 pKa = 11.84ISEE42 pKa = 4.88DD43 pKa = 3.4YY44 pKa = 10.32QATRR48 pKa = 11.84EE49 pKa = 4.04NEE51 pKa = 4.08EE52 pKa = 3.86IDD54 pKa = 4.22DD55 pKa = 3.74INYY58 pKa = 10.14EE59 pKa = 3.99KK60 pKa = 10.82EE61 pKa = 4.17KK62 pKa = 10.63EE63 pKa = 4.35SKK65 pKa = 10.26EE66 pKa = 3.95LEE68 pKa = 4.23KK69 pKa = 10.68MFTEE73 pKa = 4.59NDD75 pKa = 3.1GDD77 pKa = 4.18TNITPEE83 pKa = 3.7ILQEE87 pKa = 3.95IKK89 pKa = 10.43RR90 pKa = 11.84NHH92 pKa = 6.47HH93 pKa = 6.23LRR95 pKa = 11.84SGTIDD100 pKa = 3.5IIGQSTTSSNRR111 pKa = 11.84PYY113 pKa = 11.4LDD115 pKa = 2.98MGKK118 pKa = 10.07KK119 pKa = 9.54KK120 pKa = 10.78DD121 pKa = 4.01EE122 pKa = 4.53INLTSNDD129 pKa = 4.37CIDD132 pKa = 4.05NLNQKK137 pKa = 10.29LEE139 pKa = 4.14EE140 pKa = 4.92PNTSSSRR147 pKa = 11.84TTKK150 pKa = 10.44FDD152 pKa = 3.05VDD154 pKa = 3.49YY155 pKa = 9.58TAVNRR160 pKa = 11.84NDD162 pKa = 3.36YY163 pKa = 10.16YY164 pKa = 10.79RR165 pKa = 11.84GYY167 pKa = 10.91LKK169 pKa = 10.77ALSDD173 pKa = 3.98VNKK176 pKa = 10.5LLIDD180 pKa = 3.49NSINCKK186 pKa = 9.98FDD188 pKa = 3.48GSHH191 pKa = 6.65SDD193 pKa = 3.17NLKK196 pKa = 10.35IILFAKK202 pKa = 9.12TDD204 pKa = 3.84PEE206 pKa = 3.97ITSPISIEE214 pKa = 4.02RR215 pKa = 11.84QSQSSNKK222 pKa = 8.93TKK224 pKa = 10.37ILQQSTKK231 pKa = 10.4DD232 pKa = 3.27SEE234 pKa = 4.27YY235 pKa = 10.63TIRR238 pKa = 11.84KK239 pKa = 9.41LSDD242 pKa = 3.18QIKK245 pKa = 10.35VITFDD250 pKa = 3.61TPFGEE255 pKa = 4.19PVVIDD260 pKa = 3.44TAKK263 pKa = 10.66LIEE266 pKa = 4.46SASTYY271 pKa = 10.5LDD273 pKa = 3.22SQAAVCQLIEE283 pKa = 4.8LIEE286 pKa = 4.59SNSNEE291 pKa = 4.32SNHH294 pKa = 5.34PTILKK299 pKa = 9.79IISGIPKK306 pKa = 10.12YY307 pKa = 10.67SGLKK311 pKa = 8.22YY312 pKa = 10.33QVDD315 pKa = 3.25RR316 pKa = 11.84KK317 pKa = 10.7YY318 pKa = 10.81RR319 pKa = 11.84VV320 pKa = 2.89
MM1 pKa = 7.43SRR3 pKa = 11.84NHH5 pKa = 5.87FQKK8 pKa = 10.77LGNKK12 pKa = 9.25PDD14 pKa = 3.59IQADD18 pKa = 3.54IYY20 pKa = 11.62DD21 pKa = 4.12SANLMSQINLNAYY34 pKa = 7.05TISKK38 pKa = 9.95RR39 pKa = 11.84ISEE42 pKa = 4.88DD43 pKa = 3.4YY44 pKa = 10.32QATRR48 pKa = 11.84EE49 pKa = 4.04NEE51 pKa = 4.08EE52 pKa = 3.86IDD54 pKa = 4.22DD55 pKa = 3.74INYY58 pKa = 10.14EE59 pKa = 3.99KK60 pKa = 10.82EE61 pKa = 4.17KK62 pKa = 10.63EE63 pKa = 4.35SKK65 pKa = 10.26EE66 pKa = 3.95LEE68 pKa = 4.23KK69 pKa = 10.68MFTEE73 pKa = 4.59NDD75 pKa = 3.1GDD77 pKa = 4.18TNITPEE83 pKa = 3.7ILQEE87 pKa = 3.95IKK89 pKa = 10.43RR90 pKa = 11.84NHH92 pKa = 6.47HH93 pKa = 6.23LRR95 pKa = 11.84SGTIDD100 pKa = 3.5IIGQSTTSSNRR111 pKa = 11.84PYY113 pKa = 11.4LDD115 pKa = 2.98MGKK118 pKa = 10.07KK119 pKa = 9.54KK120 pKa = 10.78DD121 pKa = 4.01EE122 pKa = 4.53INLTSNDD129 pKa = 4.37CIDD132 pKa = 4.05NLNQKK137 pKa = 10.29LEE139 pKa = 4.14EE140 pKa = 4.92PNTSSSRR147 pKa = 11.84TTKK150 pKa = 10.44FDD152 pKa = 3.05VDD154 pKa = 3.49YY155 pKa = 9.58TAVNRR160 pKa = 11.84NDD162 pKa = 3.36YY163 pKa = 10.16YY164 pKa = 10.79RR165 pKa = 11.84GYY167 pKa = 10.91LKK169 pKa = 10.77ALSDD173 pKa = 3.98VNKK176 pKa = 10.5LLIDD180 pKa = 3.49NSINCKK186 pKa = 9.98FDD188 pKa = 3.48GSHH191 pKa = 6.65SDD193 pKa = 3.17NLKK196 pKa = 10.35IILFAKK202 pKa = 9.12TDD204 pKa = 3.84PEE206 pKa = 3.97ITSPISIEE214 pKa = 4.02RR215 pKa = 11.84QSQSSNKK222 pKa = 8.93TKK224 pKa = 10.37ILQQSTKK231 pKa = 10.4DD232 pKa = 3.27SEE234 pKa = 4.27YY235 pKa = 10.63TIRR238 pKa = 11.84KK239 pKa = 9.41LSDD242 pKa = 3.18QIKK245 pKa = 10.35VITFDD250 pKa = 3.61TPFGEE255 pKa = 4.19PVVIDD260 pKa = 3.44TAKK263 pKa = 10.66LIEE266 pKa = 4.46SASTYY271 pKa = 10.5LDD273 pKa = 3.22SQAAVCQLIEE283 pKa = 4.8LIEE286 pKa = 4.59SNSNEE291 pKa = 4.32SNHH294 pKa = 5.34PTILKK299 pKa = 9.79IISGIPKK306 pKa = 10.12YY307 pKa = 10.67SGLKK311 pKa = 8.22YY312 pKa = 10.33QVDD315 pKa = 3.25RR316 pKa = 11.84KK317 pKa = 10.7YY318 pKa = 10.81RR319 pKa = 11.84VV320 pKa = 2.89
Molecular weight: 36.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X4R286|X4R286_9RHAB Nucleocapsid protein OS=Puerto Almendras virus OX=1479613 GN=N PE=4 SV=1
MM1 pKa = 7.45FKK3 pKa = 10.64NSYY6 pKa = 10.49IMTGDD11 pKa = 3.19YY12 pKa = 10.24SVRR15 pKa = 11.84VFKK18 pKa = 10.49IRR20 pKa = 11.84YY21 pKa = 6.36EE22 pKa = 4.25SSFEE26 pKa = 4.04TLPSDD31 pKa = 4.57SDD33 pKa = 3.42IKK35 pKa = 11.19PMIEE39 pKa = 4.16HH40 pKa = 7.24FDD42 pKa = 3.6GKK44 pKa = 11.0LKK46 pKa = 10.44FLEE49 pKa = 4.3VFTFILNYY57 pKa = 9.63MVTLQIRR64 pKa = 11.84NKK66 pKa = 10.01VKK68 pKa = 9.48TAPTSIFVIKK78 pKa = 10.31FPVDD82 pKa = 3.6SRR84 pKa = 11.84WMFKK88 pKa = 10.49KK89 pKa = 10.22EE90 pKa = 3.81EE91 pKa = 4.28MYY93 pKa = 11.42SMHH96 pKa = 7.06KK97 pKa = 10.39DD98 pKa = 2.68IYY100 pKa = 10.53VEE102 pKa = 4.0NKK104 pKa = 9.62EE105 pKa = 4.0GSHH108 pKa = 6.23RR109 pKa = 11.84LRR111 pKa = 11.84VKK113 pKa = 10.61LVVDD117 pKa = 3.36LHH119 pKa = 7.01VDD121 pKa = 3.04NRR123 pKa = 11.84PRR125 pKa = 11.84DD126 pKa = 3.64RR127 pKa = 11.84LVRR130 pKa = 11.84PVMKK134 pKa = 10.48KK135 pKa = 9.74ILSKK139 pKa = 10.99LYY141 pKa = 8.93DD142 pKa = 3.44TSAEE146 pKa = 3.7RR147 pKa = 11.84KK148 pKa = 8.46MYY150 pKa = 10.57EE151 pKa = 3.53IMKK154 pKa = 10.09EE155 pKa = 4.0NNEE158 pKa = 4.28CC159 pKa = 4.12
MM1 pKa = 7.45FKK3 pKa = 10.64NSYY6 pKa = 10.49IMTGDD11 pKa = 3.19YY12 pKa = 10.24SVRR15 pKa = 11.84VFKK18 pKa = 10.49IRR20 pKa = 11.84YY21 pKa = 6.36EE22 pKa = 4.25SSFEE26 pKa = 4.04TLPSDD31 pKa = 4.57SDD33 pKa = 3.42IKK35 pKa = 11.19PMIEE39 pKa = 4.16HH40 pKa = 7.24FDD42 pKa = 3.6GKK44 pKa = 11.0LKK46 pKa = 10.44FLEE49 pKa = 4.3VFTFILNYY57 pKa = 9.63MVTLQIRR64 pKa = 11.84NKK66 pKa = 10.01VKK68 pKa = 9.48TAPTSIFVIKK78 pKa = 10.31FPVDD82 pKa = 3.6SRR84 pKa = 11.84WMFKK88 pKa = 10.49KK89 pKa = 10.22EE90 pKa = 3.81EE91 pKa = 4.28MYY93 pKa = 11.42SMHH96 pKa = 7.06KK97 pKa = 10.39DD98 pKa = 2.68IYY100 pKa = 10.53VEE102 pKa = 4.0NKK104 pKa = 9.62EE105 pKa = 4.0GSHH108 pKa = 6.23RR109 pKa = 11.84LRR111 pKa = 11.84VKK113 pKa = 10.61LVVDD117 pKa = 3.36LHH119 pKa = 7.01VDD121 pKa = 3.04NRR123 pKa = 11.84PRR125 pKa = 11.84DD126 pKa = 3.64RR127 pKa = 11.84LVRR130 pKa = 11.84PVMKK134 pKa = 10.48KK135 pKa = 9.74ILSKK139 pKa = 10.99LYY141 pKa = 8.93DD142 pKa = 3.44TSAEE146 pKa = 3.7RR147 pKa = 11.84KK148 pKa = 8.46MYY150 pKa = 10.57EE151 pKa = 3.53IMKK154 pKa = 10.09EE155 pKa = 4.0NNEE158 pKa = 4.28CC159 pKa = 4.12
Molecular weight: 19.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3522 |
80 |
2059 |
587.0 |
68.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.612 ± 0.444 | 1.562 ± 0.407 |
6.019 ± 0.4 | 6.019 ± 0.449 |
4.287 ± 0.445 | 4.373 ± 0.607 |
2.129 ± 0.382 | 9.881 ± 0.514 |
7.95 ± 0.422 | 9.256 ± 0.665 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.555 ± 0.425 | 8.092 ± 0.5 |
3.18 ± 0.332 | 2.612 ± 0.359 |
4.798 ± 0.373 | 7.411 ± 0.828 |
5.679 ± 0.513 | 4.969 ± 0.45 |
1.107 ± 0.205 | 5.508 ± 0.292 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
