
Sewage-associated circular DNA virus-16
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.36
Get precalculated fractions of proteins
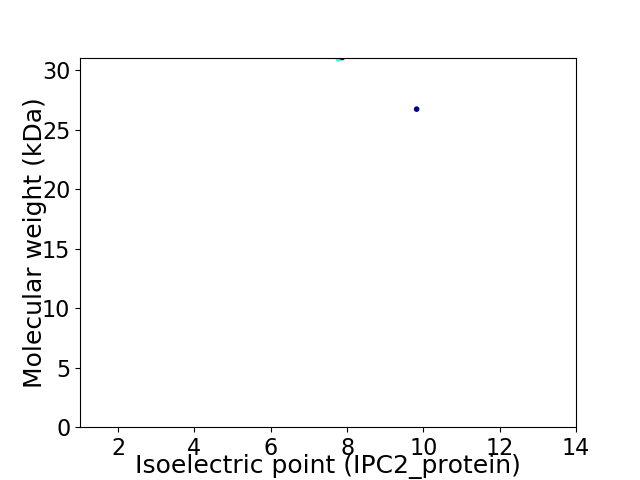
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UI40|A0A0B4UI40_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-16 OX=1592083 PE=4 SV=1
MM1 pKa = 7.88KK2 pKa = 10.45ARR4 pKa = 11.84FWILTIPHH12 pKa = 7.08ADD14 pKa = 3.53YY15 pKa = 11.44LPFLPNCCVWIRR27 pKa = 11.84GQLEE31 pKa = 3.9RR32 pKa = 11.84GSATGYY38 pKa = 9.14LHH40 pKa = 6.05WQLVVGFKK48 pKa = 8.46QQCRR52 pKa = 11.84LATLKK57 pKa = 9.97RR58 pKa = 11.84TFGDD62 pKa = 3.41SCHH65 pKa = 6.99AEE67 pKa = 4.05PTRR70 pKa = 11.84SNAADD75 pKa = 3.83DD76 pKa = 4.54YY77 pKa = 9.8VWKK80 pKa = 9.98PEE82 pKa = 3.73SRR84 pKa = 11.84IDD86 pKa = 3.42GTQFEE91 pKa = 5.15LGAKK95 pKa = 9.52PMRR98 pKa = 11.84RR99 pKa = 11.84NDD101 pKa = 3.22SRR103 pKa = 11.84DD104 pKa = 2.5WDD106 pKa = 4.59AIRR109 pKa = 11.84DD110 pKa = 3.62AAKK113 pKa = 10.49RR114 pKa = 11.84GDD116 pKa = 4.16LEE118 pKa = 4.52TVPSDD123 pKa = 3.75VFVQHH128 pKa = 5.61YY129 pKa = 9.77RR130 pKa = 11.84SLRR133 pKa = 11.84TISADD138 pKa = 3.22YY139 pKa = 10.53AEE141 pKa = 4.73PVAIEE146 pKa = 4.26RR147 pKa = 11.84TVHH150 pKa = 6.01VYY152 pKa = 9.12WGPTGVGKK160 pKa = 10.18SRR162 pKa = 11.84KK163 pKa = 7.33VWEE166 pKa = 4.28LAGEE170 pKa = 4.1DD171 pKa = 4.81AYY173 pKa = 10.19PKK175 pKa = 10.52CPNSKK180 pKa = 9.67FWDD183 pKa = 4.67GYY185 pKa = 9.57RR186 pKa = 11.84GHH188 pKa = 6.67TNVVIDD194 pKa = 4.01EE195 pKa = 4.09FRR197 pKa = 11.84GRR199 pKa = 11.84IDD201 pKa = 2.92ISHH204 pKa = 7.23ILRR207 pKa = 11.84WFDD210 pKa = 3.54RR211 pKa = 11.84YY212 pKa = 9.7PVLVEE217 pKa = 3.9IKK219 pKa = 10.53GSSTVLRR226 pKa = 11.84AQNIWITSNLDD237 pKa = 3.02PRR239 pKa = 11.84EE240 pKa = 4.15WYY242 pKa = 9.2TDD244 pKa = 3.45LDD246 pKa = 4.21EE247 pKa = 4.44LTKK250 pKa = 10.81LALLRR255 pKa = 11.84RR256 pKa = 11.84LNVIHH261 pKa = 7.14CPLALYY267 pKa = 10.08
MM1 pKa = 7.88KK2 pKa = 10.45ARR4 pKa = 11.84FWILTIPHH12 pKa = 7.08ADD14 pKa = 3.53YY15 pKa = 11.44LPFLPNCCVWIRR27 pKa = 11.84GQLEE31 pKa = 3.9RR32 pKa = 11.84GSATGYY38 pKa = 9.14LHH40 pKa = 6.05WQLVVGFKK48 pKa = 8.46QQCRR52 pKa = 11.84LATLKK57 pKa = 9.97RR58 pKa = 11.84TFGDD62 pKa = 3.41SCHH65 pKa = 6.99AEE67 pKa = 4.05PTRR70 pKa = 11.84SNAADD75 pKa = 3.83DD76 pKa = 4.54YY77 pKa = 9.8VWKK80 pKa = 9.98PEE82 pKa = 3.73SRR84 pKa = 11.84IDD86 pKa = 3.42GTQFEE91 pKa = 5.15LGAKK95 pKa = 9.52PMRR98 pKa = 11.84RR99 pKa = 11.84NDD101 pKa = 3.22SRR103 pKa = 11.84DD104 pKa = 2.5WDD106 pKa = 4.59AIRR109 pKa = 11.84DD110 pKa = 3.62AAKK113 pKa = 10.49RR114 pKa = 11.84GDD116 pKa = 4.16LEE118 pKa = 4.52TVPSDD123 pKa = 3.75VFVQHH128 pKa = 5.61YY129 pKa = 9.77RR130 pKa = 11.84SLRR133 pKa = 11.84TISADD138 pKa = 3.22YY139 pKa = 10.53AEE141 pKa = 4.73PVAIEE146 pKa = 4.26RR147 pKa = 11.84TVHH150 pKa = 6.01VYY152 pKa = 9.12WGPTGVGKK160 pKa = 10.18SRR162 pKa = 11.84KK163 pKa = 7.33VWEE166 pKa = 4.28LAGEE170 pKa = 4.1DD171 pKa = 4.81AYY173 pKa = 10.19PKK175 pKa = 10.52CPNSKK180 pKa = 9.67FWDD183 pKa = 4.67GYY185 pKa = 9.57RR186 pKa = 11.84GHH188 pKa = 6.67TNVVIDD194 pKa = 4.01EE195 pKa = 4.09FRR197 pKa = 11.84GRR199 pKa = 11.84IDD201 pKa = 2.92ISHH204 pKa = 7.23ILRR207 pKa = 11.84WFDD210 pKa = 3.54RR211 pKa = 11.84YY212 pKa = 9.7PVLVEE217 pKa = 3.9IKK219 pKa = 10.53GSSTVLRR226 pKa = 11.84AQNIWITSNLDD237 pKa = 3.02PRR239 pKa = 11.84EE240 pKa = 4.15WYY242 pKa = 9.2TDD244 pKa = 3.45LDD246 pKa = 4.21EE247 pKa = 4.44LTKK250 pKa = 10.81LALLRR255 pKa = 11.84RR256 pKa = 11.84LNVIHH261 pKa = 7.14CPLALYY267 pKa = 10.08
Molecular weight: 30.95 kDa
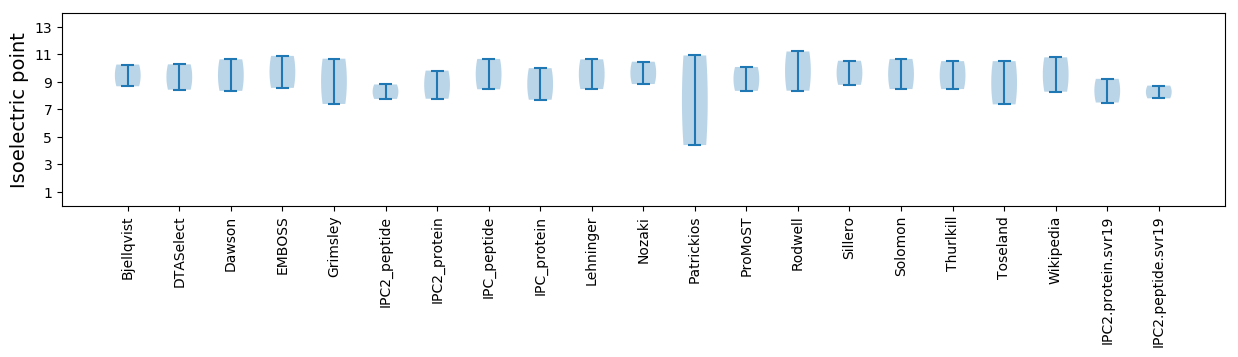
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI40|A0A0B4UI40_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-16 OX=1592083 PE=4 SV=1
MM1 pKa = 8.08PKK3 pKa = 9.97RR4 pKa = 11.84KK5 pKa = 9.65SSSKK9 pKa = 9.97RR10 pKa = 11.84SSSSKK15 pKa = 9.16RR16 pKa = 11.84RR17 pKa = 11.84SRR19 pKa = 11.84KK20 pKa = 9.58SSMSKK25 pKa = 10.18KK26 pKa = 9.74AIKK29 pKa = 10.15RR30 pKa = 11.84SVLQAVLGMSEE41 pKa = 3.71TKK43 pKa = 10.21YY44 pKa = 10.7RR45 pKa = 11.84QFADD49 pKa = 3.31EE50 pKa = 4.3NVNIFHH56 pKa = 6.65NTTTITGNAINMLQTSQGASDD77 pKa = 3.62NQRR80 pKa = 11.84IGDD83 pKa = 4.36TIRR86 pKa = 11.84AKK88 pKa = 10.57SLNIKK93 pKa = 8.38MQLFNKK99 pKa = 9.29QDD101 pKa = 3.73RR102 pKa = 11.84PNVTYY107 pKa = 10.24RR108 pKa = 11.84VLILAPLTTGVTPAPTIWKK127 pKa = 9.74GEE129 pKa = 4.28SGNKK133 pKa = 8.95MLDD136 pKa = 3.75NIDD139 pKa = 3.44TGSYY143 pKa = 9.85KK144 pKa = 10.04IIYY147 pKa = 8.92QKK149 pKa = 10.04FVKK152 pKa = 9.57WSSQTTTFSGPTTLPNYY169 pKa = 10.22SRR171 pKa = 11.84EE172 pKa = 3.88TSKK175 pKa = 10.65IISINIPLKK184 pKa = 10.53NKK186 pKa = 9.6KK187 pKa = 9.19LQYY190 pKa = 8.2STNSAVVPKK199 pKa = 10.34SSKK202 pKa = 10.92LNWTLYY208 pKa = 6.86VTPYY212 pKa = 10.89DD213 pKa = 3.71AFGTLITDD221 pKa = 4.6NIASYY226 pKa = 10.69AYY228 pKa = 10.12NARR231 pKa = 11.84LYY233 pKa = 10.94FKK235 pKa = 10.78DD236 pKa = 3.82FF237 pKa = 3.62
MM1 pKa = 8.08PKK3 pKa = 9.97RR4 pKa = 11.84KK5 pKa = 9.65SSSKK9 pKa = 9.97RR10 pKa = 11.84SSSSKK15 pKa = 9.16RR16 pKa = 11.84RR17 pKa = 11.84SRR19 pKa = 11.84KK20 pKa = 9.58SSMSKK25 pKa = 10.18KK26 pKa = 9.74AIKK29 pKa = 10.15RR30 pKa = 11.84SVLQAVLGMSEE41 pKa = 3.71TKK43 pKa = 10.21YY44 pKa = 10.7RR45 pKa = 11.84QFADD49 pKa = 3.31EE50 pKa = 4.3NVNIFHH56 pKa = 6.65NTTTITGNAINMLQTSQGASDD77 pKa = 3.62NQRR80 pKa = 11.84IGDD83 pKa = 4.36TIRR86 pKa = 11.84AKK88 pKa = 10.57SLNIKK93 pKa = 8.38MQLFNKK99 pKa = 9.29QDD101 pKa = 3.73RR102 pKa = 11.84PNVTYY107 pKa = 10.24RR108 pKa = 11.84VLILAPLTTGVTPAPTIWKK127 pKa = 9.74GEE129 pKa = 4.28SGNKK133 pKa = 8.95MLDD136 pKa = 3.75NIDD139 pKa = 3.44TGSYY143 pKa = 9.85KK144 pKa = 10.04IIYY147 pKa = 8.92QKK149 pKa = 10.04FVKK152 pKa = 9.57WSSQTTTFSGPTTLPNYY169 pKa = 10.22SRR171 pKa = 11.84EE172 pKa = 3.88TSKK175 pKa = 10.65IISINIPLKK184 pKa = 10.53NKK186 pKa = 9.6KK187 pKa = 9.19LQYY190 pKa = 8.2STNSAVVPKK199 pKa = 10.34SSKK202 pKa = 10.92LNWTLYY208 pKa = 6.86VTPYY212 pKa = 10.89DD213 pKa = 3.71AFGTLITDD221 pKa = 4.6NIASYY226 pKa = 10.69AYY228 pKa = 10.12NARR231 pKa = 11.84LYY233 pKa = 10.94FKK235 pKa = 10.78DD236 pKa = 3.82FF237 pKa = 3.62
Molecular weight: 26.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
504 |
237 |
267 |
252.0 |
28.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.349 ± 0.509 | 1.19 ± 0.702 |
5.754 ± 1.153 | 3.373 ± 0.993 |
3.373 ± 0.001 | 5.159 ± 0.554 |
1.786 ± 0.804 | 6.548 ± 0.617 |
7.143 ± 1.759 | 7.937 ± 0.699 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.587 ± 0.557 | 5.159 ± 1.436 |
4.762 ± 0.32 | 3.373 ± 0.499 |
7.54 ± 1.211 | 8.532 ± 2.184 |
7.738 ± 1.408 | 5.556 ± 0.788 |
2.778 ± 0.891 | 4.365 ± 0.163 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
