
Litorilinea aerophila
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Caldilineae; Caldilineales; Caldilineaceae; Litorilinea
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
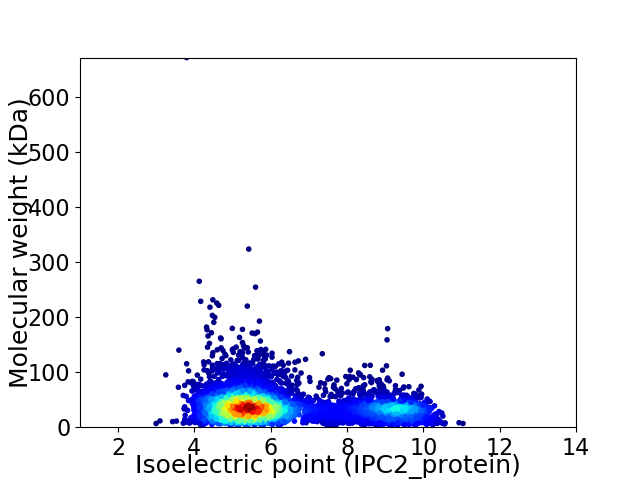
Virtual 2D-PAGE plot for 4877 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A540VGA9|A0A540VGA9_9CHLR SpoVR family protein OS=Litorilinea aerophila OX=1204385 GN=FKZ61_13430 PE=4 SV=1
MM1 pKa = 7.46TKK3 pKa = 9.95PNRR6 pKa = 11.84FIAPVCAWAFGLGLLMALVTGLTVTLSPQRR36 pKa = 11.84VTAAPLPVPHH46 pKa = 6.68FVPTLLSTTPDD57 pKa = 2.85ANARR61 pKa = 11.84HH62 pKa = 5.66VPRR65 pKa = 11.84NTAVSASFQEE75 pKa = 4.03AMNPSSVTPSTFVVHH90 pKa = 6.85GSQSGLIAGTYY101 pKa = 9.27AVSGDD106 pKa = 3.69GQTVSLTPAASLFPGEE122 pKa = 4.45RR123 pKa = 11.84VDD125 pKa = 5.97AILTTGLANTSGEE138 pKa = 4.23PLDD141 pKa = 5.38APHH144 pKa = 6.73VWQFWAQAGIATGIYY159 pKa = 9.73GFSGQPLGAANQQSYY174 pKa = 10.43AVAAGDD180 pKa = 4.01LNQDD184 pKa = 3.86GFPDD188 pKa = 4.09LVVGNAKK195 pKa = 10.52AGGHH199 pKa = 4.4NTIYY203 pKa = 10.91LNDD206 pKa = 3.81GTGSFGTISQTLPTLDD222 pKa = 3.32STRR225 pKa = 11.84SVLLVDD231 pKa = 3.95MDD233 pKa = 3.81MDD235 pKa = 4.77GDD237 pKa = 4.19LDD239 pKa = 4.25LVVGNHH245 pKa = 5.85GKK247 pKa = 9.94QNYY250 pKa = 9.52IYY252 pKa = 10.22LNNGNGTFATPGLPFGPDD270 pKa = 2.84NDD272 pKa = 4.15TTYY275 pKa = 11.67SLAAGDD281 pKa = 4.25LDD283 pKa = 4.5GNGTIDD289 pKa = 3.72LVVGNVITSPMGIYY303 pKa = 10.27FNNGNATFSSQSYY316 pKa = 7.11TLPVEE321 pKa = 4.22GQVWSVALGDD331 pKa = 3.65VDD333 pKa = 5.22GDD335 pKa = 3.84GDD337 pKa = 4.63LDD339 pKa = 4.09LAVGNYY345 pKa = 9.91SNPNQIFLNDD355 pKa = 3.3GHH357 pKa = 7.3GNFIQPGINLRR368 pKa = 11.84DD369 pKa = 3.83ATDD372 pKa = 3.48LTHH375 pKa = 7.28RR376 pKa = 11.84LVLGDD381 pKa = 3.46VDD383 pKa = 5.04GDD385 pKa = 3.87GDD387 pKa = 4.59LDD389 pKa = 4.13AVTANAGLNYY399 pKa = 10.03VYY401 pKa = 10.85YY402 pKa = 10.84NNGDD406 pKa = 3.58GTFSANRR413 pKa = 11.84HH414 pKa = 5.4PFGVSYY420 pKa = 10.68DD421 pKa = 3.6NSMGLALGDD430 pKa = 3.49VDD432 pKa = 5.64GDD434 pKa = 3.67GDD436 pKa = 4.75LDD438 pKa = 3.76VVVANDD444 pKa = 3.02VGAAKK449 pKa = 10.73NPGEE453 pKa = 4.38SGNVVYY459 pKa = 10.98LNDD462 pKa = 3.76GDD464 pKa = 4.33GTFDD468 pKa = 3.06TTAYY472 pKa = 10.74LFGGVSDD479 pKa = 4.54RR480 pKa = 11.84SQAVALADD488 pKa = 3.81FDD490 pKa = 5.26GDD492 pKa = 3.92GDD494 pKa = 4.56LDD496 pKa = 3.97LAAVNAAMAGEE507 pKa = 4.22NPVPQQSAVYY517 pKa = 8.94FNANASDD524 pKa = 3.81PAITFSMQPTSTVPGGTVRR543 pKa = 11.84YY544 pKa = 8.64FLRR547 pKa = 11.84FQNQGQAASHH557 pKa = 4.99QVIITITLPAQVTITNIEE575 pKa = 4.01TSGAPVTMSGSGQIRR590 pKa = 11.84VFQAGSLAAQASGQITVTGTLVTGLAPNTQFSATASITGSGDD632 pKa = 3.43GYY634 pKa = 11.17LLNNNASAVGQVLNGPPNAVDD655 pKa = 4.3DD656 pKa = 4.47SFTIPEE662 pKa = 4.64DD663 pKa = 3.78SPNTALSVLANDD675 pKa = 4.33SDD677 pKa = 4.65PNGDD681 pKa = 3.92PLSIYY686 pKa = 10.77AVGIPDD692 pKa = 3.43HH693 pKa = 6.6GGVVNFNASTITYY706 pKa = 9.75RR707 pKa = 11.84PAANFAGTEE716 pKa = 4.13VFTYY720 pKa = 7.43TARR723 pKa = 11.84DD724 pKa = 3.38GRR726 pKa = 11.84GGYY729 pKa = 7.9GTANVIVTVTPVNDD743 pKa = 3.88PPDD746 pKa = 3.71ARR748 pKa = 11.84NDD750 pKa = 3.55SATVDD755 pKa = 3.8EE756 pKa = 5.27DD757 pKa = 4.31SGDD760 pKa = 3.55TFIDD764 pKa = 3.35VLANDD769 pKa = 4.41TSAPDD774 pKa = 3.27TGEE777 pKa = 4.06TLTVIAVGTPSRR789 pKa = 11.84GGTVSFTPGGVTYY802 pKa = 10.56RR803 pKa = 11.84PAPNYY808 pKa = 10.61NGVEE812 pKa = 4.22TFTYY816 pKa = 9.75TISDD820 pKa = 3.94GNGGTDD826 pKa = 3.21SAQVTVTVNPVNDD839 pKa = 4.11PPDD842 pKa = 3.82ARR844 pKa = 11.84DD845 pKa = 3.43DD846 pKa = 3.83TATVDD851 pKa = 5.14EE852 pKa = 5.25DD853 pKa = 4.25SQDD856 pKa = 3.33NLINVLQNDD865 pKa = 5.2RR866 pKa = 11.84ITPDD870 pKa = 2.71VDD872 pKa = 4.14EE873 pKa = 5.11ILTVIAVGTPSQGGTATLTGGQVRR897 pKa = 11.84YY898 pKa = 7.93TPAPNFFGIEE908 pKa = 4.14TFTYY912 pKa = 9.73TISDD916 pKa = 3.94GNGGTDD922 pKa = 3.21SAQVTVTVNPVNDD935 pKa = 4.11PPDD938 pKa = 3.82ARR940 pKa = 11.84DD941 pKa = 3.43DD942 pKa = 3.83TATVDD947 pKa = 4.36EE948 pKa = 5.21DD949 pKa = 3.97SQNNLINVLANDD961 pKa = 4.19TSAPDD966 pKa = 3.27TGEE969 pKa = 4.13TLTVTAVGTPSQGGTATLTGGQVRR993 pKa = 11.84YY994 pKa = 7.93TPAPNFFGIEE1004 pKa = 4.14TFTYY1008 pKa = 9.73TISDD1012 pKa = 3.94GNGGTDD1018 pKa = 3.2SAQVSVTVNPVNDD1031 pKa = 4.11PPDD1034 pKa = 3.82ARR1036 pKa = 11.84DD1037 pKa = 3.46DD1038 pKa = 3.94TATVLQNAGATPINVLANDD1057 pKa = 4.51TYY1059 pKa = 11.53LPDD1062 pKa = 4.05APEE1065 pKa = 3.93TLTIVGVGVASAGGSVSFSVDD1086 pKa = 3.05QVFYY1090 pKa = 10.13TPPPNFIGTDD1100 pKa = 3.2TFAYY1104 pKa = 8.68TISDD1108 pKa = 3.87GNGGTDD1114 pKa = 3.21SAQVTVTVTPSSNPPANHH1132 pKa = 6.76PPAARR1137 pKa = 11.84DD1138 pKa = 3.6DD1139 pKa = 3.92VATVSKK1145 pKa = 10.58NSHH1148 pKa = 5.38SNRR1151 pKa = 11.84LDD1153 pKa = 3.22VLANDD1158 pKa = 3.65STEE1161 pKa = 4.2PDD1163 pKa = 3.05TGEE1166 pKa = 3.85QLSIIAVNDD1175 pKa = 3.77LSAGGTVTFTARR1187 pKa = 11.84YY1188 pKa = 9.3VFYY1191 pKa = 10.3TPKK1194 pKa = 10.12TDD1196 pKa = 3.75FTGVEE1201 pKa = 4.18TFAYY1205 pKa = 9.19TVDD1208 pKa = 3.87DD1209 pKa = 4.35GRR1211 pKa = 11.84GGTDD1215 pKa = 2.8WAVVTVTVTGTTGSPGNPPGDD1236 pKa = 4.41PSPGDD1241 pKa = 3.42PPGTGNRR1248 pKa = 11.84APVAVNDD1255 pKa = 4.06NYY1257 pKa = 11.04STVQGQTLTVSAAEE1271 pKa = 4.17GVLQNDD1277 pKa = 3.2QDD1279 pKa = 3.93EE1280 pKa = 5.17DD1281 pKa = 4.75GDD1283 pKa = 4.13TLTAEE1288 pKa = 4.46LVADD1292 pKa = 4.4VVQGTLSLQPDD1303 pKa = 4.04GSFVYY1308 pKa = 8.74TAPATFSGTTTFSYY1322 pKa = 10.37RR1323 pKa = 11.84AYY1325 pKa = 10.58DD1326 pKa = 3.52GQAYY1330 pKa = 10.11SGAATVVISVQAEE1343 pKa = 4.0SSGNLEE1349 pKa = 3.75LDD1351 pKa = 3.39KK1352 pKa = 11.27QIFLPVIQRR1361 pKa = 11.84ATEE1364 pKa = 3.7
MM1 pKa = 7.46TKK3 pKa = 9.95PNRR6 pKa = 11.84FIAPVCAWAFGLGLLMALVTGLTVTLSPQRR36 pKa = 11.84VTAAPLPVPHH46 pKa = 6.68FVPTLLSTTPDD57 pKa = 2.85ANARR61 pKa = 11.84HH62 pKa = 5.66VPRR65 pKa = 11.84NTAVSASFQEE75 pKa = 4.03AMNPSSVTPSTFVVHH90 pKa = 6.85GSQSGLIAGTYY101 pKa = 9.27AVSGDD106 pKa = 3.69GQTVSLTPAASLFPGEE122 pKa = 4.45RR123 pKa = 11.84VDD125 pKa = 5.97AILTTGLANTSGEE138 pKa = 4.23PLDD141 pKa = 5.38APHH144 pKa = 6.73VWQFWAQAGIATGIYY159 pKa = 9.73GFSGQPLGAANQQSYY174 pKa = 10.43AVAAGDD180 pKa = 4.01LNQDD184 pKa = 3.86GFPDD188 pKa = 4.09LVVGNAKK195 pKa = 10.52AGGHH199 pKa = 4.4NTIYY203 pKa = 10.91LNDD206 pKa = 3.81GTGSFGTISQTLPTLDD222 pKa = 3.32STRR225 pKa = 11.84SVLLVDD231 pKa = 3.95MDD233 pKa = 3.81MDD235 pKa = 4.77GDD237 pKa = 4.19LDD239 pKa = 4.25LVVGNHH245 pKa = 5.85GKK247 pKa = 9.94QNYY250 pKa = 9.52IYY252 pKa = 10.22LNNGNGTFATPGLPFGPDD270 pKa = 2.84NDD272 pKa = 4.15TTYY275 pKa = 11.67SLAAGDD281 pKa = 4.25LDD283 pKa = 4.5GNGTIDD289 pKa = 3.72LVVGNVITSPMGIYY303 pKa = 10.27FNNGNATFSSQSYY316 pKa = 7.11TLPVEE321 pKa = 4.22GQVWSVALGDD331 pKa = 3.65VDD333 pKa = 5.22GDD335 pKa = 3.84GDD337 pKa = 4.63LDD339 pKa = 4.09LAVGNYY345 pKa = 9.91SNPNQIFLNDD355 pKa = 3.3GHH357 pKa = 7.3GNFIQPGINLRR368 pKa = 11.84DD369 pKa = 3.83ATDD372 pKa = 3.48LTHH375 pKa = 7.28RR376 pKa = 11.84LVLGDD381 pKa = 3.46VDD383 pKa = 5.04GDD385 pKa = 3.87GDD387 pKa = 4.59LDD389 pKa = 4.13AVTANAGLNYY399 pKa = 10.03VYY401 pKa = 10.85YY402 pKa = 10.84NNGDD406 pKa = 3.58GTFSANRR413 pKa = 11.84HH414 pKa = 5.4PFGVSYY420 pKa = 10.68DD421 pKa = 3.6NSMGLALGDD430 pKa = 3.49VDD432 pKa = 5.64GDD434 pKa = 3.67GDD436 pKa = 4.75LDD438 pKa = 3.76VVVANDD444 pKa = 3.02VGAAKK449 pKa = 10.73NPGEE453 pKa = 4.38SGNVVYY459 pKa = 10.98LNDD462 pKa = 3.76GDD464 pKa = 4.33GTFDD468 pKa = 3.06TTAYY472 pKa = 10.74LFGGVSDD479 pKa = 4.54RR480 pKa = 11.84SQAVALADD488 pKa = 3.81FDD490 pKa = 5.26GDD492 pKa = 3.92GDD494 pKa = 4.56LDD496 pKa = 3.97LAAVNAAMAGEE507 pKa = 4.22NPVPQQSAVYY517 pKa = 8.94FNANASDD524 pKa = 3.81PAITFSMQPTSTVPGGTVRR543 pKa = 11.84YY544 pKa = 8.64FLRR547 pKa = 11.84FQNQGQAASHH557 pKa = 4.99QVIITITLPAQVTITNIEE575 pKa = 4.01TSGAPVTMSGSGQIRR590 pKa = 11.84VFQAGSLAAQASGQITVTGTLVTGLAPNTQFSATASITGSGDD632 pKa = 3.43GYY634 pKa = 11.17LLNNNASAVGQVLNGPPNAVDD655 pKa = 4.3DD656 pKa = 4.47SFTIPEE662 pKa = 4.64DD663 pKa = 3.78SPNTALSVLANDD675 pKa = 4.33SDD677 pKa = 4.65PNGDD681 pKa = 3.92PLSIYY686 pKa = 10.77AVGIPDD692 pKa = 3.43HH693 pKa = 6.6GGVVNFNASTITYY706 pKa = 9.75RR707 pKa = 11.84PAANFAGTEE716 pKa = 4.13VFTYY720 pKa = 7.43TARR723 pKa = 11.84DD724 pKa = 3.38GRR726 pKa = 11.84GGYY729 pKa = 7.9GTANVIVTVTPVNDD743 pKa = 3.88PPDD746 pKa = 3.71ARR748 pKa = 11.84NDD750 pKa = 3.55SATVDD755 pKa = 3.8EE756 pKa = 5.27DD757 pKa = 4.31SGDD760 pKa = 3.55TFIDD764 pKa = 3.35VLANDD769 pKa = 4.41TSAPDD774 pKa = 3.27TGEE777 pKa = 4.06TLTVIAVGTPSRR789 pKa = 11.84GGTVSFTPGGVTYY802 pKa = 10.56RR803 pKa = 11.84PAPNYY808 pKa = 10.61NGVEE812 pKa = 4.22TFTYY816 pKa = 9.75TISDD820 pKa = 3.94GNGGTDD826 pKa = 3.21SAQVTVTVNPVNDD839 pKa = 4.11PPDD842 pKa = 3.82ARR844 pKa = 11.84DD845 pKa = 3.43DD846 pKa = 3.83TATVDD851 pKa = 5.14EE852 pKa = 5.25DD853 pKa = 4.25SQDD856 pKa = 3.33NLINVLQNDD865 pKa = 5.2RR866 pKa = 11.84ITPDD870 pKa = 2.71VDD872 pKa = 4.14EE873 pKa = 5.11ILTVIAVGTPSQGGTATLTGGQVRR897 pKa = 11.84YY898 pKa = 7.93TPAPNFFGIEE908 pKa = 4.14TFTYY912 pKa = 9.73TISDD916 pKa = 3.94GNGGTDD922 pKa = 3.21SAQVTVTVNPVNDD935 pKa = 4.11PPDD938 pKa = 3.82ARR940 pKa = 11.84DD941 pKa = 3.43DD942 pKa = 3.83TATVDD947 pKa = 4.36EE948 pKa = 5.21DD949 pKa = 3.97SQNNLINVLANDD961 pKa = 4.19TSAPDD966 pKa = 3.27TGEE969 pKa = 4.13TLTVTAVGTPSQGGTATLTGGQVRR993 pKa = 11.84YY994 pKa = 7.93TPAPNFFGIEE1004 pKa = 4.14TFTYY1008 pKa = 9.73TISDD1012 pKa = 3.94GNGGTDD1018 pKa = 3.2SAQVSVTVNPVNDD1031 pKa = 4.11PPDD1034 pKa = 3.82ARR1036 pKa = 11.84DD1037 pKa = 3.46DD1038 pKa = 3.94TATVLQNAGATPINVLANDD1057 pKa = 4.51TYY1059 pKa = 11.53LPDD1062 pKa = 4.05APEE1065 pKa = 3.93TLTIVGVGVASAGGSVSFSVDD1086 pKa = 3.05QVFYY1090 pKa = 10.13TPPPNFIGTDD1100 pKa = 3.2TFAYY1104 pKa = 8.68TISDD1108 pKa = 3.87GNGGTDD1114 pKa = 3.21SAQVTVTVTPSSNPPANHH1132 pKa = 6.76PPAARR1137 pKa = 11.84DD1138 pKa = 3.6DD1139 pKa = 3.92VATVSKK1145 pKa = 10.58NSHH1148 pKa = 5.38SNRR1151 pKa = 11.84LDD1153 pKa = 3.22VLANDD1158 pKa = 3.65STEE1161 pKa = 4.2PDD1163 pKa = 3.05TGEE1166 pKa = 3.85QLSIIAVNDD1175 pKa = 3.77LSAGGTVTFTARR1187 pKa = 11.84YY1188 pKa = 9.3VFYY1191 pKa = 10.3TPKK1194 pKa = 10.12TDD1196 pKa = 3.75FTGVEE1201 pKa = 4.18TFAYY1205 pKa = 9.19TVDD1208 pKa = 3.87DD1209 pKa = 4.35GRR1211 pKa = 11.84GGTDD1215 pKa = 2.8WAVVTVTVTGTTGSPGNPPGDD1236 pKa = 4.41PSPGDD1241 pKa = 3.42PPGTGNRR1248 pKa = 11.84APVAVNDD1255 pKa = 4.06NYY1257 pKa = 11.04STVQGQTLTVSAAEE1271 pKa = 4.17GVLQNDD1277 pKa = 3.2QDD1279 pKa = 3.93EE1280 pKa = 5.17DD1281 pKa = 4.75GDD1283 pKa = 4.13TLTAEE1288 pKa = 4.46LVADD1292 pKa = 4.4VVQGTLSLQPDD1303 pKa = 4.04GSFVYY1308 pKa = 8.74TAPATFSGTTTFSYY1322 pKa = 10.37RR1323 pKa = 11.84AYY1325 pKa = 10.58DD1326 pKa = 3.52GQAYY1330 pKa = 10.11SGAATVVISVQAEE1343 pKa = 4.0SSGNLEE1349 pKa = 3.75LDD1351 pKa = 3.39KK1352 pKa = 11.27QIFLPVIQRR1361 pKa = 11.84ATEE1364 pKa = 3.7
Molecular weight: 140.21 kDa
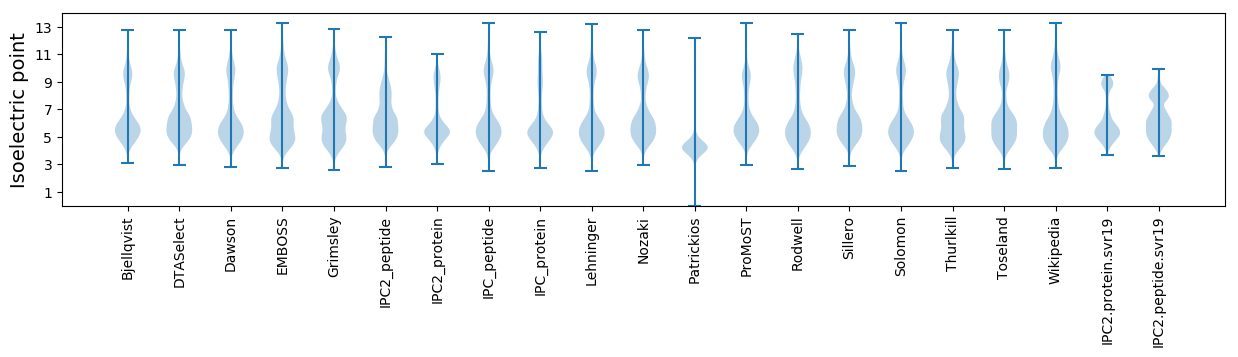
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A540V944|A0A540V944_9CHLR N-acetylmuramic acid 6-phosphate etherase OS=Litorilinea aerophila OX=1204385 GN=murQ PE=3 SV=1
MM1 pKa = 8.05PKK3 pKa = 9.16RR4 pKa = 11.84TWQPKK9 pKa = 6.52VRR11 pKa = 11.84KK12 pKa = 9.2RR13 pKa = 11.84LKK15 pKa = 8.26THH17 pKa = 5.86GFRR20 pKa = 11.84IRR22 pKa = 11.84MRR24 pKa = 11.84TPGGRR29 pKa = 11.84NVLKK33 pKa = 10.49RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84LKK38 pKa = 10.44GRR40 pKa = 11.84ARR42 pKa = 11.84LTVKK46 pKa = 9.51LTEE49 pKa = 4.24RR50 pKa = 11.84KK51 pKa = 9.26PFRR54 pKa = 11.84HH55 pKa = 6.02
MM1 pKa = 8.05PKK3 pKa = 9.16RR4 pKa = 11.84TWQPKK9 pKa = 6.52VRR11 pKa = 11.84KK12 pKa = 9.2RR13 pKa = 11.84LKK15 pKa = 8.26THH17 pKa = 5.86GFRR20 pKa = 11.84IRR22 pKa = 11.84MRR24 pKa = 11.84TPGGRR29 pKa = 11.84NVLKK33 pKa = 10.49RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84LKK38 pKa = 10.44GRR40 pKa = 11.84ARR42 pKa = 11.84LTVKK46 pKa = 9.51LTEE49 pKa = 4.24RR50 pKa = 11.84KK51 pKa = 9.26PFRR54 pKa = 11.84HH55 pKa = 6.02
Molecular weight: 6.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1775820 |
26 |
6596 |
364.1 |
40.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.031 ± 0.046 | 0.824 ± 0.011 |
5.044 ± 0.028 | 6.06 ± 0.038 |
3.528 ± 0.021 | 8.153 ± 0.028 |
2.309 ± 0.019 | 4.818 ± 0.031 |
2.051 ± 0.025 | 11.286 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.117 ± 0.017 | 2.591 ± 0.022 |
6.215 ± 0.031 | 4.454 ± 0.02 |
7.172 ± 0.039 | 4.623 ± 0.024 |
5.321 ± 0.043 | 7.67 ± 0.034 |
1.844 ± 0.019 | 2.888 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
