
Bacteroides plebeius CAG:211
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; environmental samples
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
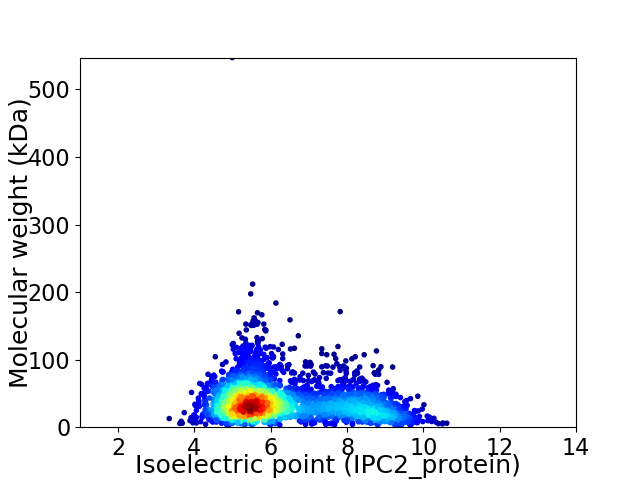
Virtual 2D-PAGE plot for 2643 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
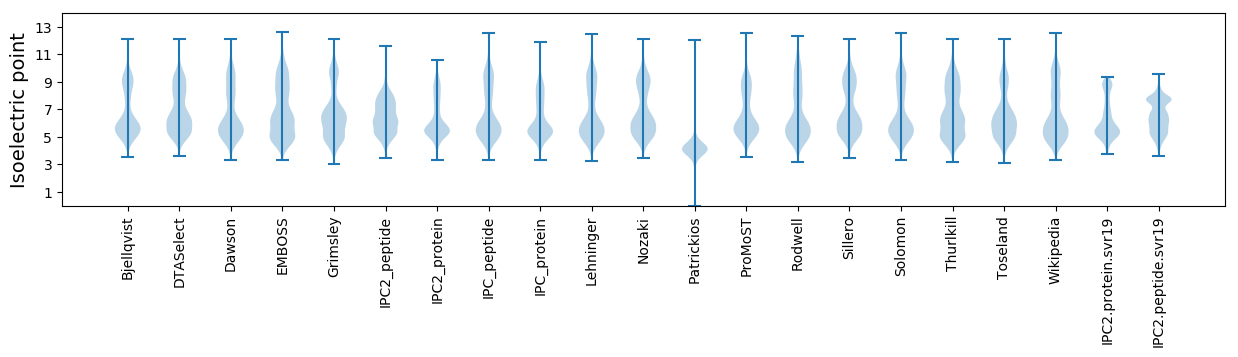
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5VRJ4|R5VRJ4_9BACE Uncharacterized protein OS=Bacteroides plebeius CAG:211 OX=1263052 GN=BN536_01077 PE=4 SV=1
MM1 pKa = 7.16KK2 pKa = 9.74TIKK5 pKa = 10.6SLFAVLFALLAFSACEE21 pKa = 3.72KK22 pKa = 10.93DD23 pKa = 3.14GDD25 pKa = 4.36KK26 pKa = 10.55IYY28 pKa = 10.82LQSLEE33 pKa = 4.71SNNLMATTDD42 pKa = 3.66RR43 pKa = 11.84VEE45 pKa = 4.44LNADD49 pKa = 4.28LAQEE53 pKa = 4.27IVVSLAWTDD62 pKa = 3.36RR63 pKa = 11.84TIQISDD69 pKa = 3.85PAVGTTVTVTNYY81 pKa = 9.91VEE83 pKa = 4.93ASLSEE88 pKa = 4.37DD89 pKa = 3.28FSTTVSKK96 pKa = 9.2TATTSLSIAFTGLEE110 pKa = 4.2LNSLASEE117 pKa = 4.23IGAVTGRR124 pKa = 11.84NNKK127 pKa = 9.58IYY129 pKa = 10.36FRR131 pKa = 11.84LAGTTGTNIPLVYY144 pKa = 10.42SNVVSIDD151 pKa = 3.37VTPYY155 pKa = 11.08KK156 pKa = 10.07MDD158 pKa = 3.62MNTGTVILSNDD169 pKa = 2.97QTEE172 pKa = 4.4NLGEE176 pKa = 4.22TGVTFYY182 pKa = 11.41SPDD185 pKa = 3.27ADD187 pKa = 4.09GIYY190 pKa = 10.35SGFISIPEE198 pKa = 3.73AWYY201 pKa = 9.4HH202 pKa = 5.88ILLKK206 pKa = 10.86EE207 pKa = 4.12GDD209 pKa = 3.55GSIWGTSGSEE219 pKa = 4.31GAFHH223 pKa = 7.23LTDD226 pKa = 4.63DD227 pKa = 5.07PNNSLQNMWFPGTAGCYY244 pKa = 8.57YY245 pKa = 10.78VIVNTQAAEE254 pKa = 3.77WSALHH259 pKa = 6.56ISSLLVSGLTDD270 pKa = 3.26EE271 pKa = 5.59SISLTLDD278 pKa = 2.88QTNNQWLATFNAEE291 pKa = 4.18TAGSRR296 pKa = 11.84TITINGKK303 pKa = 9.15GEE305 pKa = 3.94QYY307 pKa = 10.62NAEE310 pKa = 4.23TGDD313 pKa = 3.8GSGTAVDD320 pKa = 3.66IAFAANGEE328 pKa = 4.4QVSLAQTAGDD338 pKa = 3.51ITVDD342 pKa = 3.03IPQAGEE348 pKa = 3.86CTVRR352 pKa = 11.84LNLYY356 pKa = 10.25NPTNCVVSVEE366 pKa = 4.59AGASTLPGGEE376 pKa = 4.38DD377 pKa = 3.38PGEE380 pKa = 4.11EE381 pKa = 4.29TPVTLPEE388 pKa = 4.28SLDD391 pKa = 3.45VVSYY395 pKa = 9.18STGSEE400 pKa = 4.04VLLTTLYY407 pKa = 8.0PTGSGSGVYY416 pKa = 9.3TGTYY420 pKa = 8.77SGEE423 pKa = 3.86VRR425 pKa = 11.84DD426 pKa = 4.36QINIVDD432 pKa = 5.22RR433 pKa = 11.84INSVWYY439 pKa = 10.02GCAPDD444 pKa = 5.6DD445 pKa = 4.31NSQLSSQKK453 pKa = 10.68SKK455 pKa = 10.09WNIWFDD461 pKa = 3.53GSGEE465 pKa = 4.08LTLTVDD471 pKa = 4.67LTNMTWNYY479 pKa = 7.19TANN482 pKa = 3.47
MM1 pKa = 7.16KK2 pKa = 9.74TIKK5 pKa = 10.6SLFAVLFALLAFSACEE21 pKa = 3.72KK22 pKa = 10.93DD23 pKa = 3.14GDD25 pKa = 4.36KK26 pKa = 10.55IYY28 pKa = 10.82LQSLEE33 pKa = 4.71SNNLMATTDD42 pKa = 3.66RR43 pKa = 11.84VEE45 pKa = 4.44LNADD49 pKa = 4.28LAQEE53 pKa = 4.27IVVSLAWTDD62 pKa = 3.36RR63 pKa = 11.84TIQISDD69 pKa = 3.85PAVGTTVTVTNYY81 pKa = 9.91VEE83 pKa = 4.93ASLSEE88 pKa = 4.37DD89 pKa = 3.28FSTTVSKK96 pKa = 9.2TATTSLSIAFTGLEE110 pKa = 4.2LNSLASEE117 pKa = 4.23IGAVTGRR124 pKa = 11.84NNKK127 pKa = 9.58IYY129 pKa = 10.36FRR131 pKa = 11.84LAGTTGTNIPLVYY144 pKa = 10.42SNVVSIDD151 pKa = 3.37VTPYY155 pKa = 11.08KK156 pKa = 10.07MDD158 pKa = 3.62MNTGTVILSNDD169 pKa = 2.97QTEE172 pKa = 4.4NLGEE176 pKa = 4.22TGVTFYY182 pKa = 11.41SPDD185 pKa = 3.27ADD187 pKa = 4.09GIYY190 pKa = 10.35SGFISIPEE198 pKa = 3.73AWYY201 pKa = 9.4HH202 pKa = 5.88ILLKK206 pKa = 10.86EE207 pKa = 4.12GDD209 pKa = 3.55GSIWGTSGSEE219 pKa = 4.31GAFHH223 pKa = 7.23LTDD226 pKa = 4.63DD227 pKa = 5.07PNNSLQNMWFPGTAGCYY244 pKa = 8.57YY245 pKa = 10.78VIVNTQAAEE254 pKa = 3.77WSALHH259 pKa = 6.56ISSLLVSGLTDD270 pKa = 3.26EE271 pKa = 5.59SISLTLDD278 pKa = 2.88QTNNQWLATFNAEE291 pKa = 4.18TAGSRR296 pKa = 11.84TITINGKK303 pKa = 9.15GEE305 pKa = 3.94QYY307 pKa = 10.62NAEE310 pKa = 4.23TGDD313 pKa = 3.8GSGTAVDD320 pKa = 3.66IAFAANGEE328 pKa = 4.4QVSLAQTAGDD338 pKa = 3.51ITVDD342 pKa = 3.03IPQAGEE348 pKa = 3.86CTVRR352 pKa = 11.84LNLYY356 pKa = 10.25NPTNCVVSVEE366 pKa = 4.59AGASTLPGGEE376 pKa = 4.38DD377 pKa = 3.38PGEE380 pKa = 4.11EE381 pKa = 4.29TPVTLPEE388 pKa = 4.28SLDD391 pKa = 3.45VVSYY395 pKa = 9.18STGSEE400 pKa = 4.04VLLTTLYY407 pKa = 8.0PTGSGSGVYY416 pKa = 9.3TGTYY420 pKa = 8.77SGEE423 pKa = 3.86VRR425 pKa = 11.84DD426 pKa = 4.36QINIVDD432 pKa = 5.22RR433 pKa = 11.84INSVWYY439 pKa = 10.02GCAPDD444 pKa = 5.6DD445 pKa = 4.31NSQLSSQKK453 pKa = 10.68SKK455 pKa = 10.09WNIWFDD461 pKa = 3.53GSGEE465 pKa = 4.08LTLTVDD471 pKa = 4.67LTNMTWNYY479 pKa = 7.19TANN482 pKa = 3.47
Molecular weight: 51.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5V6A6|R5V6A6_9BACE Dihydroorotate oxidase OS=Bacteroides plebeius CAG:211 OX=1263052 GN=BN536_01014 PE=4 SV=1
MM1 pKa = 7.72CGFFLANFPCWIDD14 pKa = 4.5FYY16 pKa = 10.85PLQCALFSLFGKK28 pKa = 8.53KK29 pKa = 6.51TTQKK33 pKa = 9.93FADD36 pKa = 3.72YY37 pKa = 10.72KK38 pKa = 10.18FSCTFATAIEE48 pKa = 4.25NEE50 pKa = 4.58TIGRR54 pKa = 11.84LAQLVQSICLTSRR67 pKa = 11.84GSAVRR72 pKa = 11.84IRR74 pKa = 11.84QRR76 pKa = 11.84PLNEE80 pKa = 3.74FLVNHH85 pKa = 6.54CFFFLGRR92 pKa = 11.84LAQLVQSICLTSRR105 pKa = 11.84GSAVRR110 pKa = 11.84IRR112 pKa = 11.84QRR114 pKa = 11.84PHH116 pKa = 6.86RR117 pKa = 11.84NIIVNFPPIGRR128 pKa = 11.84LAQLVQSICLTSRR141 pKa = 11.84GSAVRR146 pKa = 11.84IRR148 pKa = 11.84QRR150 pKa = 11.84PLQASQFCGAFFFF163 pKa = 5.44
MM1 pKa = 7.72CGFFLANFPCWIDD14 pKa = 4.5FYY16 pKa = 10.85PLQCALFSLFGKK28 pKa = 8.53KK29 pKa = 6.51TTQKK33 pKa = 9.93FADD36 pKa = 3.72YY37 pKa = 10.72KK38 pKa = 10.18FSCTFATAIEE48 pKa = 4.25NEE50 pKa = 4.58TIGRR54 pKa = 11.84LAQLVQSICLTSRR67 pKa = 11.84GSAVRR72 pKa = 11.84IRR74 pKa = 11.84QRR76 pKa = 11.84PLNEE80 pKa = 3.74FLVNHH85 pKa = 6.54CFFFLGRR92 pKa = 11.84LAQLVQSICLTSRR105 pKa = 11.84GSAVRR110 pKa = 11.84IRR112 pKa = 11.84QRR114 pKa = 11.84PHH116 pKa = 6.86RR117 pKa = 11.84NIIVNFPPIGRR128 pKa = 11.84LAQLVQSICLTSRR141 pKa = 11.84GSAVRR146 pKa = 11.84IRR148 pKa = 11.84QRR150 pKa = 11.84PLQASQFCGAFFFF163 pKa = 5.44
Molecular weight: 18.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
961016 |
29 |
4860 |
363.6 |
41.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.211 ± 0.047 | 1.343 ± 0.017 |
5.345 ± 0.033 | 6.84 ± 0.047 |
4.541 ± 0.029 | 7.031 ± 0.042 |
2.015 ± 0.02 | 6.336 ± 0.045 |
6.42 ± 0.041 | 9.29 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.857 ± 0.023 | 4.773 ± 0.04 |
3.769 ± 0.022 | 3.693 ± 0.031 |
4.678 ± 0.031 | 5.95 ± 0.038 |
5.525 ± 0.033 | 6.708 ± 0.038 |
1.296 ± 0.019 | 4.379 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
