
Bacillus sp. JCM 19045
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; unclassified Bacillus (in: Bacteria)
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
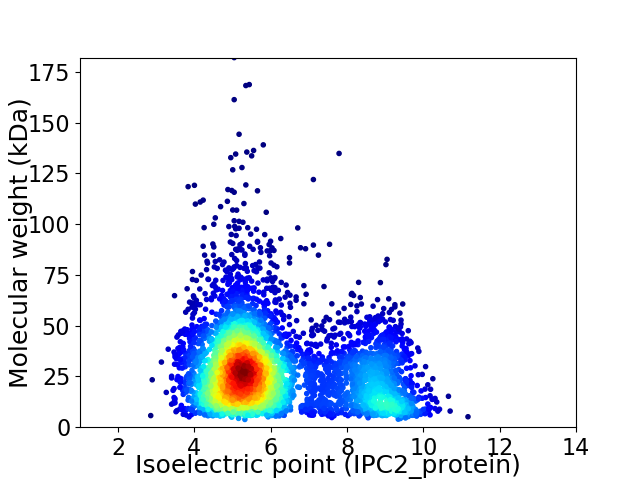
Virtual 2D-PAGE plot for 4678 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W7ZC55|W7ZC55_9BACI Response regulator aspartate phosphatase OS=Bacillus sp. JCM 19045 OX=1460639 GN=JCM19045_3380 PE=3 SV=1
MM1 pKa = 7.37KK2 pKa = 10.53KK3 pKa = 10.35SMEE6 pKa = 4.15YY7 pKa = 11.1GLVSTMAIALLAGCNQGSSGGEE29 pKa = 3.69DD30 pKa = 3.25VVEE33 pKa = 4.66LSFSAWGNPAEE44 pKa = 4.04LEE46 pKa = 4.45VYY48 pKa = 10.2NRR50 pKa = 11.84AVDD53 pKa = 3.81AFNEE57 pKa = 4.25EE58 pKa = 3.61QDD60 pKa = 4.15EE61 pKa = 4.42IQVTLTGIPNDD72 pKa = 3.79NYY74 pKa = 10.55FQTLTTRR81 pKa = 11.84LQGGQAPDD89 pKa = 3.18VFYY92 pKa = 11.31VGSEE96 pKa = 4.01NMSTLIANGTIEE108 pKa = 4.19PMSDD112 pKa = 3.53FMGTDD117 pKa = 3.05EE118 pKa = 6.08SYY120 pKa = 10.21VQEE123 pKa = 5.02SEE125 pKa = 4.11FTDD128 pKa = 5.63DD129 pKa = 2.61IWGPSKK135 pKa = 10.58QDD137 pKa = 3.43EE138 pKa = 4.56IVYY141 pKa = 9.53GLPVDD146 pKa = 4.31CNPILMYY153 pKa = 10.6YY154 pKa = 10.61NKK156 pKa = 9.55TLLEE160 pKa = 4.39DD161 pKa = 3.95AGIEE165 pKa = 4.19SPQTYY170 pKa = 9.61FEE172 pKa = 4.13QGDD175 pKa = 4.1WNWDD179 pKa = 3.17TFAEE183 pKa = 4.29ITGKK187 pKa = 10.5LKK189 pKa = 10.4EE190 pKa = 4.62AGNHH194 pKa = 6.06GYY196 pKa = 8.91VQEE199 pKa = 4.58SGHH202 pKa = 6.29GNLINWVWTNGGNFATDD219 pKa = 3.55DD220 pKa = 4.93AIVADD225 pKa = 4.19TDD227 pKa = 3.79EE228 pKa = 4.5LTLEE232 pKa = 3.94AFEE235 pKa = 4.88YY236 pKa = 9.9VDD238 pKa = 4.74EE239 pKa = 4.4MVQAGNFTYY248 pKa = 10.57AGTMPDD254 pKa = 3.34GQGIEE259 pKa = 4.35GMFMTNSVGFVSAGRR274 pKa = 11.84WLTPMFLQTSLDD286 pKa = 3.3FDD288 pKa = 5.0YY289 pKa = 11.26IPFPSNTGEE298 pKa = 4.42PIEE301 pKa = 4.25TATIATAYY309 pKa = 7.96MAANSKK315 pKa = 10.38SEE317 pKa = 4.15HH318 pKa = 6.27VEE320 pKa = 3.71EE321 pKa = 4.18AQKK324 pKa = 10.76FISYY328 pKa = 6.46YY329 pKa = 10.46TSMEE333 pKa = 4.04GQRR336 pKa = 11.84VRR338 pKa = 11.84LDD340 pKa = 3.55GEE342 pKa = 4.63GNAVPSVEE350 pKa = 4.54GIDD353 pKa = 5.7DD354 pKa = 3.48IVLNQDD360 pKa = 3.29RR361 pKa = 11.84PEE363 pKa = 4.52HH364 pKa = 5.43GQYY367 pKa = 11.25LLDD370 pKa = 3.88ARR372 pKa = 11.84SVGRR376 pKa = 11.84VINYY380 pKa = 7.6EE381 pKa = 3.93FTSPGLNNDD390 pKa = 3.0LDD392 pKa = 4.55EE393 pKa = 6.08IYY395 pKa = 11.08EE396 pKa = 4.08LMLLGDD402 pKa = 3.82MTGEE406 pKa = 4.03EE407 pKa = 4.55ALDD410 pKa = 3.63EE411 pKa = 4.43ASRR414 pKa = 11.84VAKK417 pKa = 10.39AALDD421 pKa = 3.78EE422 pKa = 4.4
MM1 pKa = 7.37KK2 pKa = 10.53KK3 pKa = 10.35SMEE6 pKa = 4.15YY7 pKa = 11.1GLVSTMAIALLAGCNQGSSGGEE29 pKa = 3.69DD30 pKa = 3.25VVEE33 pKa = 4.66LSFSAWGNPAEE44 pKa = 4.04LEE46 pKa = 4.45VYY48 pKa = 10.2NRR50 pKa = 11.84AVDD53 pKa = 3.81AFNEE57 pKa = 4.25EE58 pKa = 3.61QDD60 pKa = 4.15EE61 pKa = 4.42IQVTLTGIPNDD72 pKa = 3.79NYY74 pKa = 10.55FQTLTTRR81 pKa = 11.84LQGGQAPDD89 pKa = 3.18VFYY92 pKa = 11.31VGSEE96 pKa = 4.01NMSTLIANGTIEE108 pKa = 4.19PMSDD112 pKa = 3.53FMGTDD117 pKa = 3.05EE118 pKa = 6.08SYY120 pKa = 10.21VQEE123 pKa = 5.02SEE125 pKa = 4.11FTDD128 pKa = 5.63DD129 pKa = 2.61IWGPSKK135 pKa = 10.58QDD137 pKa = 3.43EE138 pKa = 4.56IVYY141 pKa = 9.53GLPVDD146 pKa = 4.31CNPILMYY153 pKa = 10.6YY154 pKa = 10.61NKK156 pKa = 9.55TLLEE160 pKa = 4.39DD161 pKa = 3.95AGIEE165 pKa = 4.19SPQTYY170 pKa = 9.61FEE172 pKa = 4.13QGDD175 pKa = 4.1WNWDD179 pKa = 3.17TFAEE183 pKa = 4.29ITGKK187 pKa = 10.5LKK189 pKa = 10.4EE190 pKa = 4.62AGNHH194 pKa = 6.06GYY196 pKa = 8.91VQEE199 pKa = 4.58SGHH202 pKa = 6.29GNLINWVWTNGGNFATDD219 pKa = 3.55DD220 pKa = 4.93AIVADD225 pKa = 4.19TDD227 pKa = 3.79EE228 pKa = 4.5LTLEE232 pKa = 3.94AFEE235 pKa = 4.88YY236 pKa = 9.9VDD238 pKa = 4.74EE239 pKa = 4.4MVQAGNFTYY248 pKa = 10.57AGTMPDD254 pKa = 3.34GQGIEE259 pKa = 4.35GMFMTNSVGFVSAGRR274 pKa = 11.84WLTPMFLQTSLDD286 pKa = 3.3FDD288 pKa = 5.0YY289 pKa = 11.26IPFPSNTGEE298 pKa = 4.42PIEE301 pKa = 4.25TATIATAYY309 pKa = 7.96MAANSKK315 pKa = 10.38SEE317 pKa = 4.15HH318 pKa = 6.27VEE320 pKa = 3.71EE321 pKa = 4.18AQKK324 pKa = 10.76FISYY328 pKa = 6.46YY329 pKa = 10.46TSMEE333 pKa = 4.04GQRR336 pKa = 11.84VRR338 pKa = 11.84LDD340 pKa = 3.55GEE342 pKa = 4.63GNAVPSVEE350 pKa = 4.54GIDD353 pKa = 5.7DD354 pKa = 3.48IVLNQDD360 pKa = 3.29RR361 pKa = 11.84PEE363 pKa = 4.52HH364 pKa = 5.43GQYY367 pKa = 11.25LLDD370 pKa = 3.88ARR372 pKa = 11.84SVGRR376 pKa = 11.84VINYY380 pKa = 7.6EE381 pKa = 3.93FTSPGLNNDD390 pKa = 3.0LDD392 pKa = 4.55EE393 pKa = 6.08IYY395 pKa = 11.08EE396 pKa = 4.08LMLLGDD402 pKa = 3.82MTGEE406 pKa = 4.03EE407 pKa = 4.55ALDD410 pKa = 3.63EE411 pKa = 4.43ASRR414 pKa = 11.84VAKK417 pKa = 10.39AALDD421 pKa = 3.78EE422 pKa = 4.4
Molecular weight: 46.49 kDa
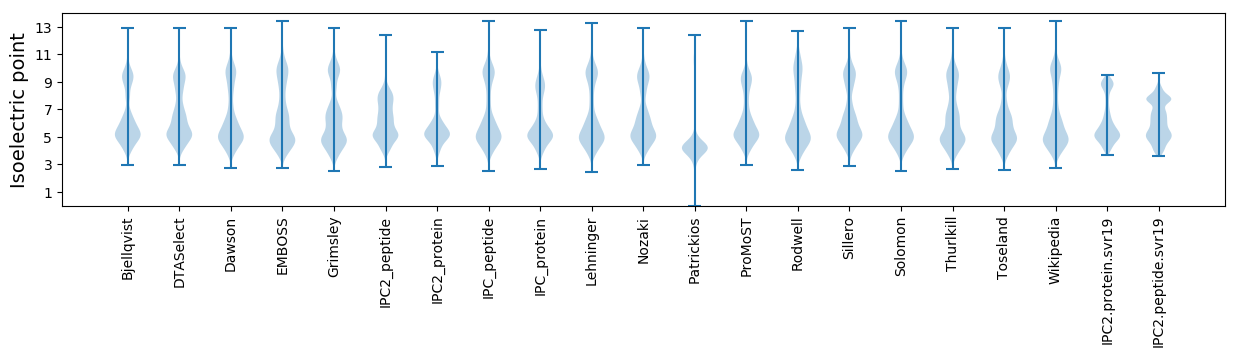
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W7Z1D5|W7Z1D5_9BACI GMP synthase (glutamine-hydrolyzing) OS=Bacillus sp. JCM 19045 OX=1460639 GN=JCM19045_4088 PE=4 SV=1
MM1 pKa = 7.61GPTFQPNNRR10 pKa = 11.84KK11 pKa = 9.34RR12 pKa = 11.84KK13 pKa = 8.16KK14 pKa = 9.1NHH16 pKa = 4.73GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 8.82VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.39GRR39 pKa = 11.84KK40 pKa = 8.52VLSAA44 pKa = 4.05
MM1 pKa = 7.61GPTFQPNNRR10 pKa = 11.84KK11 pKa = 9.34RR12 pKa = 11.84KK13 pKa = 8.16KK14 pKa = 9.1NHH16 pKa = 4.73GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37NGRR28 pKa = 11.84KK29 pKa = 8.82VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.39GRR39 pKa = 11.84KK40 pKa = 8.52VLSAA44 pKa = 4.05
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1234247 |
37 |
1642 |
263.8 |
29.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.78 ± 0.042 | 0.703 ± 0.011 |
5.084 ± 0.035 | 7.462 ± 0.053 |
4.582 ± 0.029 | 6.807 ± 0.041 |
2.22 ± 0.019 | 7.056 ± 0.037 |
5.609 ± 0.036 | 10.24 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.756 ± 0.02 | 3.993 ± 0.026 |
3.579 ± 0.019 | 4.264 ± 0.025 |
4.27 ± 0.024 | 6.2 ± 0.027 |
5.825 ± 0.032 | 7.083 ± 0.032 |
1.086 ± 0.016 | 3.4 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
