
HIV-1 CRF03_AB
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Orthoretrovirinae; Lentivirus; Human immunodeficiency virus 1; HIV-1 recombinants; HIV-1 circulating recombinant forms
Average proteome isoelectric point is 7.32
Get precalculated fractions of proteins
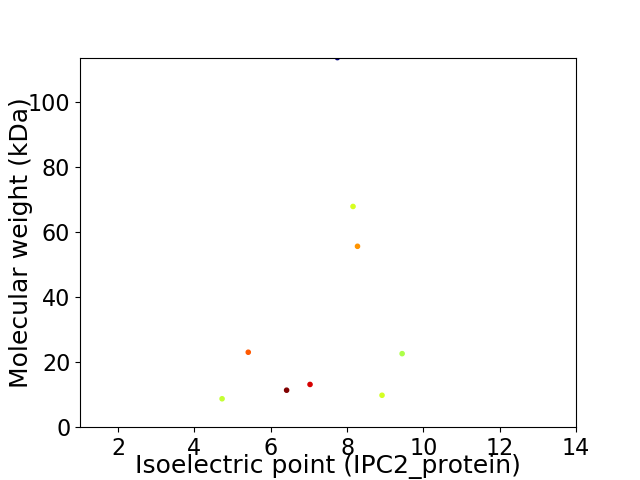
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9Q6W0|Q9Q6W0_9HIV1 Protein Rev OS=HIV-1 CRF03_AB OX=540995 GN=rev PE=3 SV=1
MM1 pKa = 7.19QSLAIAAIVALVVVGIIAIVVGSIVFIEE29 pKa = 3.91YY30 pKa = 10.41RR31 pKa = 11.84KK32 pKa = 9.66ILRR35 pKa = 11.84QRR37 pKa = 11.84KK38 pKa = 8.02IDD40 pKa = 3.69RR41 pKa = 11.84LIDD44 pKa = 4.43RR45 pKa = 11.84IRR47 pKa = 11.84EE48 pKa = 3.87RR49 pKa = 11.84AEE51 pKa = 3.95DD52 pKa = 3.58SGNEE56 pKa = 4.05SEE58 pKa = 5.01GDD60 pKa = 3.47QEE62 pKa = 5.53ALMEE66 pKa = 4.45MGHH69 pKa = 6.54LVPWDD74 pKa = 3.81ADD76 pKa = 3.76DD77 pKa = 4.23LL78 pKa = 4.39
MM1 pKa = 7.19QSLAIAAIVALVVVGIIAIVVGSIVFIEE29 pKa = 3.91YY30 pKa = 10.41RR31 pKa = 11.84KK32 pKa = 9.66ILRR35 pKa = 11.84QRR37 pKa = 11.84KK38 pKa = 8.02IDD40 pKa = 3.69RR41 pKa = 11.84LIDD44 pKa = 4.43RR45 pKa = 11.84IRR47 pKa = 11.84EE48 pKa = 3.87RR49 pKa = 11.84AEE51 pKa = 3.95DD52 pKa = 3.58SGNEE56 pKa = 4.05SEE58 pKa = 5.01GDD60 pKa = 3.47QEE62 pKa = 5.53ALMEE66 pKa = 4.45MGHH69 pKa = 6.54LVPWDD74 pKa = 3.81ADD76 pKa = 3.76DD77 pKa = 4.23LL78 pKa = 4.39
Molecular weight: 8.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9Q6W3|Q9Q6W3_9HIV1 Exoribonuclease H (Fragment) OS=HIV-1 CRF03_AB OX=540995 GN=pol PE=3 SV=1
MM1 pKa = 7.3EE2 pKa = 5.21NRR4 pKa = 11.84WQVMIVWQVDD14 pKa = 3.04RR15 pKa = 11.84MRR17 pKa = 11.84IRR19 pKa = 11.84TWNSLVKK26 pKa = 9.66HH27 pKa = 6.66HH28 pKa = 7.07IYY30 pKa = 10.07ISKK33 pKa = 10.1KK34 pKa = 9.13ARR36 pKa = 11.84GWVYY40 pKa = 9.39KK41 pKa = 10.13HH42 pKa = 6.55HH43 pKa = 6.38YY44 pKa = 8.7EE45 pKa = 4.24SRR47 pKa = 11.84NPRR50 pKa = 11.84ISSEE54 pKa = 3.9VHH56 pKa = 5.92IPLGDD61 pKa = 3.41AKK63 pKa = 10.86LVIKK67 pKa = 7.92TYY69 pKa = 9.48WGLHH73 pKa = 4.19TGEE76 pKa = 4.83RR77 pKa = 11.84DD78 pKa = 2.79WHH80 pKa = 6.28LGQGASIEE88 pKa = 4.16WRR90 pKa = 11.84KK91 pKa = 9.44EE92 pKa = 3.85RR93 pKa = 11.84YY94 pKa = 7.6STQVDD99 pKa = 3.82PNLADD104 pKa = 3.66QLIHH108 pKa = 7.2LYY110 pKa = 10.94YY111 pKa = 10.17FDD113 pKa = 5.17CFSEE117 pKa = 4.36SAIRR121 pKa = 11.84NAILGHH127 pKa = 6.04RR128 pKa = 11.84VSPSCEE134 pKa = 3.33YY135 pKa = 10.5RR136 pKa = 11.84AGHH139 pKa = 5.98NKK141 pKa = 9.62VGSLQYY147 pKa = 10.69LALAALRR154 pKa = 11.84TPKK157 pKa = 10.17KK158 pKa = 9.75IKK160 pKa = 10.14PPLPSVTKK168 pKa = 9.62LTEE171 pKa = 4.23DD172 pKa = 3.16RR173 pKa = 11.84WNKK176 pKa = 7.51PQRR179 pKa = 11.84TKK181 pKa = 10.81DD182 pKa = 3.24HH183 pKa = 6.83RR184 pKa = 11.84GSHH187 pKa = 5.16TMSGHH192 pKa = 5.5
MM1 pKa = 7.3EE2 pKa = 5.21NRR4 pKa = 11.84WQVMIVWQVDD14 pKa = 3.04RR15 pKa = 11.84MRR17 pKa = 11.84IRR19 pKa = 11.84TWNSLVKK26 pKa = 9.66HH27 pKa = 6.66HH28 pKa = 7.07IYY30 pKa = 10.07ISKK33 pKa = 10.1KK34 pKa = 9.13ARR36 pKa = 11.84GWVYY40 pKa = 9.39KK41 pKa = 10.13HH42 pKa = 6.55HH43 pKa = 6.38YY44 pKa = 8.7EE45 pKa = 4.24SRR47 pKa = 11.84NPRR50 pKa = 11.84ISSEE54 pKa = 3.9VHH56 pKa = 5.92IPLGDD61 pKa = 3.41AKK63 pKa = 10.86LVIKK67 pKa = 7.92TYY69 pKa = 9.48WGLHH73 pKa = 4.19TGEE76 pKa = 4.83RR77 pKa = 11.84DD78 pKa = 2.79WHH80 pKa = 6.28LGQGASIEE88 pKa = 4.16WRR90 pKa = 11.84KK91 pKa = 9.44EE92 pKa = 3.85RR93 pKa = 11.84YY94 pKa = 7.6STQVDD99 pKa = 3.82PNLADD104 pKa = 3.66QLIHH108 pKa = 7.2LYY110 pKa = 10.94YY111 pKa = 10.17FDD113 pKa = 5.17CFSEE117 pKa = 4.36SAIRR121 pKa = 11.84NAILGHH127 pKa = 6.04RR128 pKa = 11.84VSPSCEE134 pKa = 3.33YY135 pKa = 10.5RR136 pKa = 11.84AGHH139 pKa = 5.98NKK141 pKa = 9.62VGSLQYY147 pKa = 10.69LALAALRR154 pKa = 11.84TPKK157 pKa = 10.17KK158 pKa = 9.75IKK160 pKa = 10.14PPLPSVTKK168 pKa = 9.62LTEE171 pKa = 4.23DD172 pKa = 3.16RR173 pKa = 11.84WNKK176 pKa = 7.51PQRR179 pKa = 11.84TKK181 pKa = 10.81DD182 pKa = 3.24HH183 pKa = 6.83RR184 pKa = 11.84GSHH187 pKa = 5.16TMSGHH192 pKa = 5.5
Molecular weight: 22.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2877 |
78 |
1003 |
319.7 |
36.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.083 ± 0.399 | 1.981 ± 0.486 |
4.414 ± 0.404 | 6.882 ± 0.534 |
2.711 ± 0.342 | 7.334 ± 0.27 |
2.225 ± 0.634 | 6.813 ± 0.73 |
7.369 ± 0.842 | 7.994 ± 0.284 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.051 ± 0.367 | 4.101 ± 0.705 |
5.666 ± 0.605 | 5.909 ± 0.507 |
5.77 ± 1.137 | 5.492 ± 0.686 |
5.874 ± 0.79 | 6.152 ± 0.571 |
2.537 ± 0.24 | 2.642 ± 0.315 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
