
Chaetoceros protobacilladnavirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Baphyvirales; Bacilladnaviridae; Protobacilladnavirus
Average proteome isoelectric point is 8.36
Get precalculated fractions of proteins
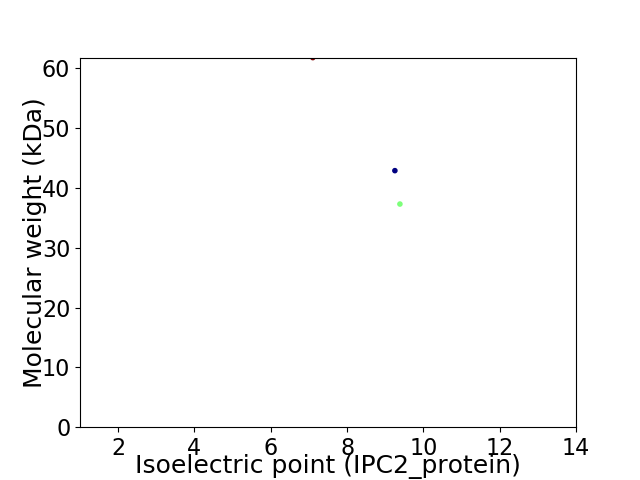
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
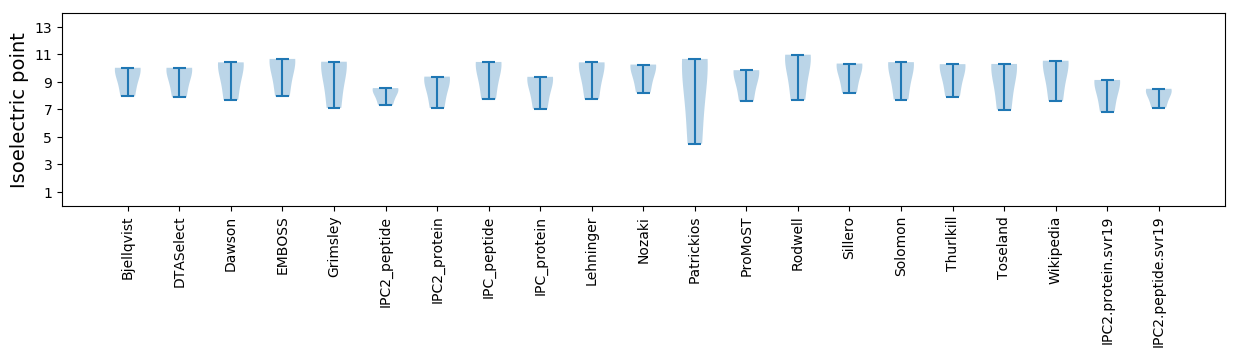
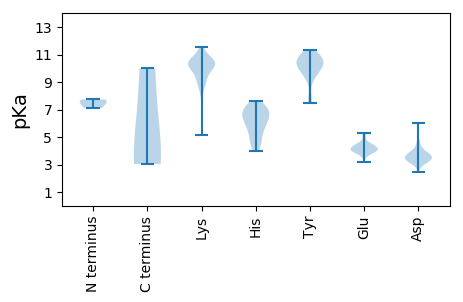
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E9RFF0|E9RFF0_9VIRU Isoform of E9RFF1 Hypothetical protein protein OS=Chaetoceros protobacilladnavirus 3 OX=2170142 GN=hypothetical protein PE=4 SV=1
MM1 pKa = 7.1RR2 pKa = 11.84QKK4 pKa = 10.65SINEE8 pKa = 3.85ILCNGLPQQEE18 pKa = 4.66LLPSEE23 pKa = 5.1SVFSDD28 pKa = 3.31SAEE31 pKa = 3.95SRR33 pKa = 11.84LLKK36 pKa = 10.44EE37 pKa = 4.05WVNDD41 pKa = 4.17FSDD44 pKa = 3.76SSPIGLDD51 pKa = 3.16LGSLDD56 pKa = 3.95TVPEE60 pKa = 3.91FDD62 pKa = 3.51VASSKK67 pKa = 10.45RR68 pKa = 11.84ASSLTTPGTLNPIQPLSRR86 pKa = 11.84PLVRR90 pKa = 11.84STAVDD95 pKa = 3.39MTPSEE100 pKa = 4.52TEE102 pKa = 4.11DD103 pKa = 3.69EE104 pKa = 4.49LSGTHH109 pKa = 6.46QDD111 pKa = 3.28PCSKK115 pKa = 10.44KK116 pKa = 10.26DD117 pKa = 3.37QLLSRR122 pKa = 11.84FILTFFPEE130 pKa = 4.3TKK132 pKa = 9.17EE133 pKa = 4.05DD134 pKa = 3.12RR135 pKa = 11.84WMEE138 pKa = 4.12PDD140 pKa = 3.27TYY142 pKa = 11.0FPGAVEE148 pKa = 5.09LFHH151 pKa = 6.87VWTGQYY157 pKa = 9.59EE158 pKa = 4.28EE159 pKa = 5.26CPTTKK164 pKa = 10.03RR165 pKa = 11.84LHH167 pKa = 4.92AHH169 pKa = 6.98IYY171 pKa = 10.89VEE173 pKa = 4.03MLTNRR178 pKa = 11.84RR179 pKa = 11.84KK180 pKa = 10.3RR181 pKa = 11.84FNAFHH186 pKa = 5.4KK187 pKa = 10.17HH188 pKa = 4.0IRR190 pKa = 11.84KK191 pKa = 8.34FHH193 pKa = 6.54KK194 pKa = 10.22GVQIRR199 pKa = 11.84SARR202 pKa = 11.84RR203 pKa = 11.84CSVKK207 pKa = 10.09QRR209 pKa = 11.84QCAINYY215 pKa = 9.41VLDD218 pKa = 3.69DD219 pKa = 4.04RR220 pKa = 11.84KK221 pKa = 10.44RR222 pKa = 11.84LEE224 pKa = 4.53GSDD227 pKa = 3.43TYY229 pKa = 9.63TWRR232 pKa = 11.84HH233 pKa = 4.29SHH235 pKa = 7.25FEE237 pKa = 3.89PKK239 pKa = 10.0YY240 pKa = 9.42DD241 pKa = 4.2SKK243 pKa = 11.47FEE245 pKa = 4.06KK246 pKa = 9.2TKK248 pKa = 10.81EE249 pKa = 3.85MVKK252 pKa = 10.28RR253 pKa = 11.84GEE255 pKa = 4.02DD256 pKa = 3.49SEE258 pKa = 4.46TQRR261 pKa = 11.84VYY263 pKa = 10.66IEE265 pKa = 4.26SKK267 pKa = 9.53PRR269 pKa = 11.84MWTWDD274 pKa = 3.62QIVHH278 pKa = 5.6EE279 pKa = 4.36TDD281 pKa = 2.96EE282 pKa = 4.63SKK284 pKa = 11.18KK285 pKa = 10.89LLFNCSWGNKK295 pKa = 6.06YY296 pKa = 10.14HH297 pKa = 7.02AGRR300 pKa = 11.84VAEE303 pKa = 4.19VSRR306 pKa = 11.84RR307 pKa = 11.84TIQQVIILYY316 pKa = 10.05GAGGTGKK323 pKa = 7.42TTTVQSWDD331 pKa = 3.14ARR333 pKa = 11.84EE334 pKa = 4.18NEE336 pKa = 4.37DD337 pKa = 3.5PRR339 pKa = 11.84EE340 pKa = 3.7RR341 pKa = 11.84YY342 pKa = 9.12YY343 pKa = 11.01RR344 pKa = 11.84RR345 pKa = 11.84NPDD348 pKa = 4.7DD349 pKa = 3.61GAFWGGGRR357 pKa = 11.84TAYY360 pKa = 10.09RR361 pKa = 11.84GQRR364 pKa = 11.84IVHH367 pKa = 5.34YY368 pKa = 10.43EE369 pKa = 3.82EE370 pKa = 4.32FTGQEE375 pKa = 3.72AFCRR379 pKa = 11.84LKK381 pKa = 10.54EE382 pKa = 3.9VCDD385 pKa = 3.78IGKK388 pKa = 9.21HH389 pKa = 4.64GPPVNVKK396 pKa = 9.74MGGTDD401 pKa = 3.55LNHH404 pKa = 7.08EE405 pKa = 4.09IVIFTTNVHH414 pKa = 5.7PAGWFHH420 pKa = 7.11KK421 pKa = 10.41LWANDD426 pKa = 3.48PKK428 pKa = 10.7QFHH431 pKa = 6.23PFWRR435 pKa = 11.84RR436 pKa = 11.84VTKK439 pKa = 10.49VMFYY443 pKa = 10.28PSHH446 pKa = 6.97RR447 pKa = 11.84PDD449 pKa = 3.4GQLNCPDD456 pKa = 4.89DD457 pKa = 3.91EE458 pKa = 5.02HH459 pKa = 7.02PPHH462 pKa = 7.63VVDD465 pKa = 5.44QTEE468 pKa = 4.03DD469 pKa = 3.13WQSFKK474 pKa = 11.29GDD476 pKa = 3.37YY477 pKa = 10.49AKK479 pKa = 10.8CLNHH483 pKa = 7.35AEE485 pKa = 4.42KK486 pKa = 10.36FWPLKK491 pKa = 10.49LEE493 pKa = 4.36PEE495 pKa = 4.31STPFSRR501 pKa = 11.84DD502 pKa = 3.06FGHH505 pKa = 7.03SRR507 pKa = 11.84GQTEE511 pKa = 3.66QSTPFFHH518 pKa = 6.42YY519 pKa = 9.8CKK521 pKa = 10.2SGRR524 pKa = 11.84DD525 pKa = 3.52PTKK528 pKa = 10.64RR529 pKa = 11.84SII531 pKa = 3.65
MM1 pKa = 7.1RR2 pKa = 11.84QKK4 pKa = 10.65SINEE8 pKa = 3.85ILCNGLPQQEE18 pKa = 4.66LLPSEE23 pKa = 5.1SVFSDD28 pKa = 3.31SAEE31 pKa = 3.95SRR33 pKa = 11.84LLKK36 pKa = 10.44EE37 pKa = 4.05WVNDD41 pKa = 4.17FSDD44 pKa = 3.76SSPIGLDD51 pKa = 3.16LGSLDD56 pKa = 3.95TVPEE60 pKa = 3.91FDD62 pKa = 3.51VASSKK67 pKa = 10.45RR68 pKa = 11.84ASSLTTPGTLNPIQPLSRR86 pKa = 11.84PLVRR90 pKa = 11.84STAVDD95 pKa = 3.39MTPSEE100 pKa = 4.52TEE102 pKa = 4.11DD103 pKa = 3.69EE104 pKa = 4.49LSGTHH109 pKa = 6.46QDD111 pKa = 3.28PCSKK115 pKa = 10.44KK116 pKa = 10.26DD117 pKa = 3.37QLLSRR122 pKa = 11.84FILTFFPEE130 pKa = 4.3TKK132 pKa = 9.17EE133 pKa = 4.05DD134 pKa = 3.12RR135 pKa = 11.84WMEE138 pKa = 4.12PDD140 pKa = 3.27TYY142 pKa = 11.0FPGAVEE148 pKa = 5.09LFHH151 pKa = 6.87VWTGQYY157 pKa = 9.59EE158 pKa = 4.28EE159 pKa = 5.26CPTTKK164 pKa = 10.03RR165 pKa = 11.84LHH167 pKa = 4.92AHH169 pKa = 6.98IYY171 pKa = 10.89VEE173 pKa = 4.03MLTNRR178 pKa = 11.84RR179 pKa = 11.84KK180 pKa = 10.3RR181 pKa = 11.84FNAFHH186 pKa = 5.4KK187 pKa = 10.17HH188 pKa = 4.0IRR190 pKa = 11.84KK191 pKa = 8.34FHH193 pKa = 6.54KK194 pKa = 10.22GVQIRR199 pKa = 11.84SARR202 pKa = 11.84RR203 pKa = 11.84CSVKK207 pKa = 10.09QRR209 pKa = 11.84QCAINYY215 pKa = 9.41VLDD218 pKa = 3.69DD219 pKa = 4.04RR220 pKa = 11.84KK221 pKa = 10.44RR222 pKa = 11.84LEE224 pKa = 4.53GSDD227 pKa = 3.43TYY229 pKa = 9.63TWRR232 pKa = 11.84HH233 pKa = 4.29SHH235 pKa = 7.25FEE237 pKa = 3.89PKK239 pKa = 10.0YY240 pKa = 9.42DD241 pKa = 4.2SKK243 pKa = 11.47FEE245 pKa = 4.06KK246 pKa = 9.2TKK248 pKa = 10.81EE249 pKa = 3.85MVKK252 pKa = 10.28RR253 pKa = 11.84GEE255 pKa = 4.02DD256 pKa = 3.49SEE258 pKa = 4.46TQRR261 pKa = 11.84VYY263 pKa = 10.66IEE265 pKa = 4.26SKK267 pKa = 9.53PRR269 pKa = 11.84MWTWDD274 pKa = 3.62QIVHH278 pKa = 5.6EE279 pKa = 4.36TDD281 pKa = 2.96EE282 pKa = 4.63SKK284 pKa = 11.18KK285 pKa = 10.89LLFNCSWGNKK295 pKa = 6.06YY296 pKa = 10.14HH297 pKa = 7.02AGRR300 pKa = 11.84VAEE303 pKa = 4.19VSRR306 pKa = 11.84RR307 pKa = 11.84TIQQVIILYY316 pKa = 10.05GAGGTGKK323 pKa = 7.42TTTVQSWDD331 pKa = 3.14ARR333 pKa = 11.84EE334 pKa = 4.18NEE336 pKa = 4.37DD337 pKa = 3.5PRR339 pKa = 11.84EE340 pKa = 3.7RR341 pKa = 11.84YY342 pKa = 9.12YY343 pKa = 11.01RR344 pKa = 11.84RR345 pKa = 11.84NPDD348 pKa = 4.7DD349 pKa = 3.61GAFWGGGRR357 pKa = 11.84TAYY360 pKa = 10.09RR361 pKa = 11.84GQRR364 pKa = 11.84IVHH367 pKa = 5.34YY368 pKa = 10.43EE369 pKa = 3.82EE370 pKa = 4.32FTGQEE375 pKa = 3.72AFCRR379 pKa = 11.84LKK381 pKa = 10.54EE382 pKa = 3.9VCDD385 pKa = 3.78IGKK388 pKa = 9.21HH389 pKa = 4.64GPPVNVKK396 pKa = 9.74MGGTDD401 pKa = 3.55LNHH404 pKa = 7.08EE405 pKa = 4.09IVIFTTNVHH414 pKa = 5.7PAGWFHH420 pKa = 7.11KK421 pKa = 10.41LWANDD426 pKa = 3.48PKK428 pKa = 10.7QFHH431 pKa = 6.23PFWRR435 pKa = 11.84RR436 pKa = 11.84VTKK439 pKa = 10.49VMFYY443 pKa = 10.28PSHH446 pKa = 6.97RR447 pKa = 11.84PDD449 pKa = 3.4GQLNCPDD456 pKa = 4.89DD457 pKa = 3.91EE458 pKa = 5.02HH459 pKa = 7.02PPHH462 pKa = 7.63VVDD465 pKa = 5.44QTEE468 pKa = 4.03DD469 pKa = 3.13WQSFKK474 pKa = 11.29GDD476 pKa = 3.37YY477 pKa = 10.49AKK479 pKa = 10.8CLNHH483 pKa = 7.35AEE485 pKa = 4.42KK486 pKa = 10.36FWPLKK491 pKa = 10.49LEE493 pKa = 4.36PEE495 pKa = 4.31STPFSRR501 pKa = 11.84DD502 pKa = 3.06FGHH505 pKa = 7.03SRR507 pKa = 11.84GQTEE511 pKa = 3.66QSTPFFHH518 pKa = 6.42YY519 pKa = 9.8CKK521 pKa = 10.2SGRR524 pKa = 11.84DD525 pKa = 3.52PTKK528 pKa = 10.64RR529 pKa = 11.84SII531 pKa = 3.65
Molecular weight: 61.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E9RFF2|E9RFF2_9VIRU Replication-associated protein OS=Chaetoceros protobacilladnavirus 3 OX=2170142 PE=4 SV=1
MM1 pKa = 7.73AGRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 8.99QRR8 pKa = 11.84RR9 pKa = 11.84SRR11 pKa = 11.84APRR14 pKa = 11.84RR15 pKa = 11.84KK16 pKa = 8.62RR17 pKa = 11.84TTGRR21 pKa = 11.84KK22 pKa = 5.15TLKK25 pKa = 10.35RR26 pKa = 11.84KK27 pKa = 9.68ADD29 pKa = 3.55SRR31 pKa = 11.84ATPAKK36 pKa = 9.15STRR39 pKa = 11.84KK40 pKa = 8.0RR41 pKa = 11.84TKK43 pKa = 9.07TVSGAAKK50 pKa = 9.06HH51 pKa = 5.57HH52 pKa = 5.91AVRR55 pKa = 11.84VKK57 pKa = 10.18PFSNATTQPKK67 pKa = 10.07IPDD70 pKa = 3.75GLLTSSLSRR79 pKa = 11.84RR80 pKa = 11.84LQNVVGVRR88 pKa = 11.84NGNSPSVHH96 pKa = 6.34AGSDD100 pKa = 3.51VMHH103 pKa = 6.04VVIAPTLGVPVMIANSAEE121 pKa = 4.17GVLKK125 pKa = 10.65RR126 pKa = 11.84PGLSQEE132 pKa = 3.91SSFIGFPGQTVGFEE146 pKa = 4.1NLIEE150 pKa = 4.38STGVPTWPPTIPTGQKK166 pKa = 10.19LEE168 pKa = 4.23NKK170 pKa = 9.99GGFVLWRR177 pKa = 11.84IISQGLRR184 pKa = 11.84IDD186 pKa = 4.25LANSDD191 pKa = 4.54EE192 pKa = 4.75EE193 pKa = 4.21NDD195 pKa = 3.53GWFEE199 pKa = 3.94ACRR202 pKa = 11.84FNWRR206 pKa = 11.84NVPRR210 pKa = 11.84DD211 pKa = 3.34VCMTPLDD218 pKa = 4.04GSTTTNSIGIAPNPLWLEE236 pKa = 3.85EE237 pKa = 4.08VGYY240 pKa = 10.79GMAMVEE246 pKa = 3.98QPGYY250 pKa = 10.92KK251 pKa = 9.7SGLLKK256 pKa = 10.44DD257 pKa = 3.38IKK259 pKa = 10.47KK260 pKa = 10.68AEE262 pKa = 4.23FMLHH266 pKa = 6.39PRR268 pKa = 11.84TTTHH272 pKa = 7.37DD273 pKa = 3.53PTLIDD278 pKa = 3.59PFEE281 pKa = 4.69YY282 pKa = 10.27GGSMTSSGGIDD293 pKa = 3.17NVYY296 pKa = 10.17YY297 pKa = 10.3PSDD300 pKa = 3.78NVSGNAVRR308 pKa = 11.84FRR310 pKa = 11.84DD311 pKa = 3.49MGVDD315 pKa = 3.92QNMDD319 pKa = 3.53WIYY322 pKa = 10.88IRR324 pKa = 11.84LHH326 pKa = 5.82CRR328 pKa = 11.84PNNGTSSLGSNFLFNVIQNVEE349 pKa = 4.12VAFNPSSDD357 pKa = 3.18FAAFQTINKK366 pKa = 9.38ADD368 pKa = 3.85TKK370 pKa = 10.47TKK372 pKa = 9.93MVADD376 pKa = 4.47GLNNNPDD383 pKa = 3.35VFNGRR388 pKa = 11.84RR389 pKa = 11.84KK390 pKa = 10.0
MM1 pKa = 7.73AGRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 8.99QRR8 pKa = 11.84RR9 pKa = 11.84SRR11 pKa = 11.84APRR14 pKa = 11.84RR15 pKa = 11.84KK16 pKa = 8.62RR17 pKa = 11.84TTGRR21 pKa = 11.84KK22 pKa = 5.15TLKK25 pKa = 10.35RR26 pKa = 11.84KK27 pKa = 9.68ADD29 pKa = 3.55SRR31 pKa = 11.84ATPAKK36 pKa = 9.15STRR39 pKa = 11.84KK40 pKa = 8.0RR41 pKa = 11.84TKK43 pKa = 9.07TVSGAAKK50 pKa = 9.06HH51 pKa = 5.57HH52 pKa = 5.91AVRR55 pKa = 11.84VKK57 pKa = 10.18PFSNATTQPKK67 pKa = 10.07IPDD70 pKa = 3.75GLLTSSLSRR79 pKa = 11.84RR80 pKa = 11.84LQNVVGVRR88 pKa = 11.84NGNSPSVHH96 pKa = 6.34AGSDD100 pKa = 3.51VMHH103 pKa = 6.04VVIAPTLGVPVMIANSAEE121 pKa = 4.17GVLKK125 pKa = 10.65RR126 pKa = 11.84PGLSQEE132 pKa = 3.91SSFIGFPGQTVGFEE146 pKa = 4.1NLIEE150 pKa = 4.38STGVPTWPPTIPTGQKK166 pKa = 10.19LEE168 pKa = 4.23NKK170 pKa = 9.99GGFVLWRR177 pKa = 11.84IISQGLRR184 pKa = 11.84IDD186 pKa = 4.25LANSDD191 pKa = 4.54EE192 pKa = 4.75EE193 pKa = 4.21NDD195 pKa = 3.53GWFEE199 pKa = 3.94ACRR202 pKa = 11.84FNWRR206 pKa = 11.84NVPRR210 pKa = 11.84DD211 pKa = 3.34VCMTPLDD218 pKa = 4.04GSTTTNSIGIAPNPLWLEE236 pKa = 3.85EE237 pKa = 4.08VGYY240 pKa = 10.79GMAMVEE246 pKa = 3.98QPGYY250 pKa = 10.92KK251 pKa = 9.7SGLLKK256 pKa = 10.44DD257 pKa = 3.38IKK259 pKa = 10.47KK260 pKa = 10.68AEE262 pKa = 4.23FMLHH266 pKa = 6.39PRR268 pKa = 11.84TTTHH272 pKa = 7.37DD273 pKa = 3.53PTLIDD278 pKa = 3.59PFEE281 pKa = 4.69YY282 pKa = 10.27GGSMTSSGGIDD293 pKa = 3.17NVYY296 pKa = 10.17YY297 pKa = 10.3PSDD300 pKa = 3.78NVSGNAVRR308 pKa = 11.84FRR310 pKa = 11.84DD311 pKa = 3.49MGVDD315 pKa = 3.92QNMDD319 pKa = 3.53WIYY322 pKa = 10.88IRR324 pKa = 11.84LHH326 pKa = 5.82CRR328 pKa = 11.84PNNGTSSLGSNFLFNVIQNVEE349 pKa = 4.12VAFNPSSDD357 pKa = 3.18FAAFQTINKK366 pKa = 9.38ADD368 pKa = 3.85TKK370 pKa = 10.47TKK372 pKa = 9.93MVADD376 pKa = 4.47GLNNNPDD383 pKa = 3.35VFNGRR388 pKa = 11.84RR389 pKa = 11.84KK390 pKa = 10.0
Molecular weight: 42.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1252 |
331 |
531 |
417.3 |
47.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.99 ± 1.353 | 1.198 ± 0.475 |
5.751 ± 0.347 | 5.911 ± 0.957 |
4.073 ± 0.755 | 7.268 ± 0.72 |
3.195 ± 0.634 | 4.473 ± 0.529 |
7.508 ± 1.187 | 6.47 ± 0.364 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.917 ± 0.323 | 4.633 ± 0.955 |
5.751 ± 0.59 | 3.834 ± 0.367 |
7.907 ± 0.181 | 7.348 ± 0.249 |
6.15 ± 0.916 | 6.15 ± 0.475 |
1.837 ± 0.445 | 2.636 ± 0.387 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
