
Pseudoxanthomonas wuyuanensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Pseudoxanthomonas
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
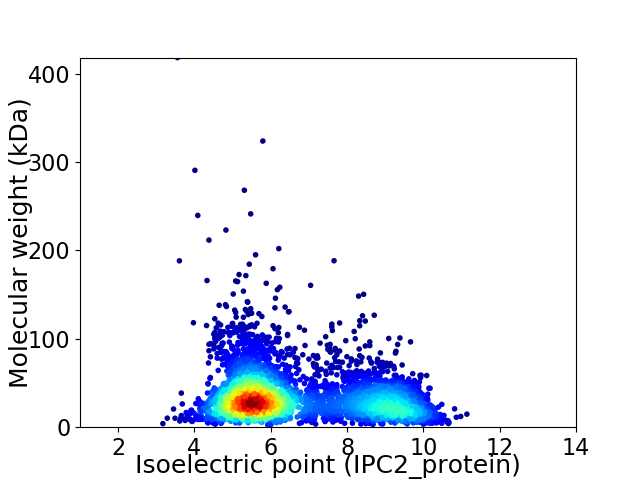
Virtual 2D-PAGE plot for 3989 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A286D655|A0A286D655_9GAMM Transcriptional regulator AraC family OS=Pseudoxanthomonas wuyuanensis OX=1073196 GN=SAMN06296416_10395 PE=4 SV=1
SS1 pKa = 6.03VGSVGAEE8 pKa = 3.35RR9 pKa = 11.84TITNVAAGRR18 pKa = 11.84ISGSSTDD25 pKa = 4.69AINGSQLFATNQAIAAIAASTPVHH49 pKa = 5.88YY50 pKa = 10.42FSVNDD55 pKa = 4.71GGTQQANYY63 pKa = 10.08LNDD66 pKa = 3.61GATAPFAIAIGSNATATGSYY86 pKa = 7.95GTAVGEE92 pKa = 4.24GATAIGDD99 pKa = 3.48WATAYY104 pKa = 10.73GNDD107 pKa = 3.68ANAGTTTATAIGAGTSATATFAFGAGFQATASGVSSTAVGLHH149 pKa = 6.25ANASGEE155 pKa = 4.16SGIAVGTDD163 pKa = 2.9AASTGTYY170 pKa = 10.17AIAIGRR176 pKa = 11.84NASATGSHH184 pKa = 5.36GTAVGEE190 pKa = 4.2GATAIGDD197 pKa = 3.48WATAYY202 pKa = 10.75GNGANASTTTATAIGGGTSATATFAFGAGYY232 pKa = 9.67QATAGGIASTAVGLHH247 pKa = 6.15ANATGEE253 pKa = 4.07SSIAIGTDD261 pKa = 3.03AVADD265 pKa = 3.7SDD267 pKa = 3.91EE268 pKa = 4.14AVAIGRR274 pKa = 11.84GSVATGGRR282 pKa = 11.84AVAIGSGNVANGDD295 pKa = 3.61GAVAIGDD302 pKa = 4.13PNTATGNGAIASGLDD317 pKa = 3.17NTATGNGSVAMGNTNMVGGGGQAVSTPGTAAQGAVGIGYY356 pKa = 9.49QNTVIGQGSVALGSTSSALAAGAVAFGDD384 pKa = 3.51TAVANNAGDD393 pKa = 3.63VALGSGSVTDD403 pKa = 3.76TAVATASTVIDD414 pKa = 3.59GVTYY418 pKa = 10.85NFAGTTPTSTVSVGAVGSEE437 pKa = 3.58RR438 pKa = 11.84TITNVAAGRR447 pKa = 11.84ISDD450 pKa = 3.88TSTDD454 pKa = 4.24AINGSQLYY462 pKa = 8.24ATNQAIEE469 pKa = 4.26AVATTASAGWNLSAQGANATNVAPDD494 pKa = 3.44EE495 pKa = 4.33TVDD498 pKa = 5.09LNNSDD503 pKa = 3.65GNIVVSKK510 pKa = 9.28TATDD514 pKa = 3.82DD515 pKa = 4.84DD516 pKa = 4.4VTFDD520 pKa = 4.12LAADD524 pKa = 3.52ITVDD528 pKa = 3.32SVTAGGTVLNTNGLTIAGGPSVTTTGINAGGLVISNVAPGVAGTDD573 pKa = 3.37AVNVDD578 pKa = 3.45QLTSVSTAGLNFTGNDD594 pKa = 3.08NTAGDD599 pKa = 3.62VHH601 pKa = 7.43RR602 pKa = 11.84DD603 pKa = 3.27LGQTLAIQGGASTAGTYY620 pKa = 10.39SGGNLRR626 pKa = 11.84TVTDD630 pKa = 4.13PVTGAIQLQMADD642 pKa = 3.75SPVFGATTVNAGGTGIISGVTAGVADD668 pKa = 4.0TDD670 pKa = 4.0AVNVSQLNAIGDD682 pKa = 3.75IANAGWNISAQGANATNVAPDD703 pKa = 3.44EE704 pKa = 4.33TVDD707 pKa = 5.09LNNSDD712 pKa = 3.65GNIVVSKK719 pKa = 9.28TATDD723 pKa = 3.82DD724 pKa = 4.84DD725 pKa = 4.4VTFDD729 pKa = 4.12LAADD733 pKa = 3.52ITVDD737 pKa = 3.78SVTAGNSVLNTSGLTVDD754 pKa = 5.19DD755 pKa = 4.84GVNSTSVGSTGLVITGGPSVTTTGIDD781 pKa = 3.07AGSLVISNVAPGVAGTDD798 pKa = 3.37AVNVDD803 pKa = 3.45QLTSVSTAGLNFTGNDD819 pKa = 3.08NTAGDD824 pKa = 3.62VHH826 pKa = 7.43RR827 pKa = 11.84DD828 pKa = 3.27LGQTLAIRR836 pKa = 11.84GGASTAGTYY845 pKa = 10.39SGGNLRR851 pKa = 11.84TVTDD855 pKa = 4.01PATGAIQLQMADD867 pKa = 3.56SPVFGGTTVNAGGTGIISGVTAGVADD893 pKa = 4.0TDD895 pKa = 4.0AVNVSQLNAIGDD907 pKa = 3.74IATAGWNLSAQGANATNVAPDD928 pKa = 3.44EE929 pKa = 4.33TVDD932 pKa = 5.09LNNSDD937 pKa = 3.65GNIVVSKK944 pKa = 10.71NATDD948 pKa = 4.25DD949 pKa = 4.77DD950 pKa = 4.54VTFDD954 pKa = 4.04LADD957 pKa = 5.61DD958 pKa = 3.76ITVDD962 pKa = 3.41SVTAGGTVLNSNGLTIAGGPSVTTTGINAGGLVISNVAPGVAGTDD1007 pKa = 3.37AVNVDD1012 pKa = 3.25QLTAVGNIANAGWNITDD1029 pKa = 3.84GTNSGNIAPNEE1040 pKa = 4.2TLTVAAGSNATVVYY1054 pKa = 10.29DD1055 pKa = 4.38DD1056 pKa = 3.78STGTMTVGVVADD1068 pKa = 4.12PSFNSITVGDD1078 pKa = 3.89TVINDD1083 pKa = 3.41GSISFTSGGPSITNIGIDD1101 pKa = 3.75AGNTTITNVAAAVNNTDD1118 pKa = 3.52AVNLEE1123 pKa = 4.05QVTNLVEE1130 pKa = 4.49GATTRR1135 pKa = 11.84YY1136 pKa = 10.47YY1137 pKa = 11.01SVNDD1141 pKa = 3.38NGVQGGNYY1149 pKa = 9.72NNDD1152 pKa = 3.02GATGINAMAAGVGASSAGAQGVALGNNASVITDD1185 pKa = 3.7GGVALGSGSLSDD1197 pKa = 4.39RR1198 pKa = 11.84VVAPQSGGIIAGTGLIPYY1216 pKa = 7.62NTTDD1220 pKa = 3.11KK1221 pKa = 10.43TLLGAVSVGTDD1232 pKa = 2.96TSYY1235 pKa = 11.36RR1236 pKa = 11.84QITNVADD1243 pKa = 3.7GTEE1246 pKa = 4.21DD1247 pKa = 3.21QDD1249 pKa = 5.01AVTLRR1254 pKa = 11.84QLRR1257 pKa = 11.84NGLSSFAVTPTMYY1270 pKa = 10.49FHH1272 pKa = 7.45ANSTAPDD1279 pKa = 3.48SLAIGAEE1286 pKa = 4.33SVAIGPNTTVNGDD1299 pKa = 3.18NGIGMGNGALVQQTAPGGIAIGQDD1323 pKa = 3.5TIVNFADD1330 pKa = 4.6GVALGTNAVTNGIQAMALGAGATANEE1356 pKa = 4.54PGSVALGAGSVTAVAVATPSTTIDD1380 pKa = 3.1GVTYY1384 pKa = 10.32NYY1386 pKa = 10.9AGTAPTSTVSVGAAGTEE1403 pKa = 3.85RR1404 pKa = 11.84TITHH1408 pKa = 7.08LAAGRR1413 pKa = 11.84ISADD1417 pKa = 2.96STDD1420 pKa = 3.76AVNGSQLFAAHH1431 pKa = 6.1QSIEE1435 pKa = 4.1NLSTRR1440 pKa = 11.84IDD1442 pKa = 3.46ASATHH1447 pKa = 6.45YY1448 pKa = 11.15YY1449 pKa = 10.24SVNDD1453 pKa = 3.53NGVVGGNYY1461 pKa = 10.14NNDD1464 pKa = 3.12GATGINSLATGVDD1477 pKa = 3.7SAASGDD1483 pKa = 3.49ASVAIGFGATAADD1496 pKa = 3.99ANSVALGSNATTAAAVGTASIVIGGTTYY1524 pKa = 11.23NFAGTAPVGTVSVGDD1539 pKa = 3.55VGAEE1543 pKa = 3.55RR1544 pKa = 11.84TITNVAAGRR1553 pKa = 11.84IGADD1557 pKa = 2.83STDD1560 pKa = 3.41AVNGSQLHH1568 pKa = 5.69ATNQAIANLSNTVNASTAHH1587 pKa = 5.95YY1588 pKa = 10.49YY1589 pKa = 10.36SVNDD1593 pKa = 3.9GGVQGGNYY1601 pKa = 9.87NNDD1604 pKa = 3.26GASGVNAVASGVNAAASGQGSVAVGANSNAAGAGSVAVGSGSQSTVAGGVAIGADD1659 pKa = 3.88SVSSTAAGVAGYY1671 pKa = 10.28VPAGATAGQAAAIAATTSTSGAVSVGDD1698 pKa = 3.99ASSGLYY1704 pKa = 9.79RR1705 pKa = 11.84QITGVAAGTAASDD1718 pKa = 3.81AVNVAQLQAVQNQLNATATHH1738 pKa = 6.57YY1739 pKa = 11.18YY1740 pKa = 9.73SVNDD1744 pKa = 3.92GGVQGGNYY1752 pKa = 9.8NNDD1755 pKa = 3.03GATGVNSMAGGVNSTASGSNSVALGANSNASAANSAALGSGSQATHH1801 pKa = 5.76VNSVAIGNGSATTVGAQTGYY1821 pKa = 9.17NAAYY1825 pKa = 9.9VGSSNSTGEE1834 pKa = 3.89VNMGGRR1840 pKa = 11.84TITGVAPGIAGTDD1853 pKa = 3.24AVNVNQLNAGVNHH1866 pKa = 7.21AVNQANQYY1874 pKa = 9.04TDD1876 pKa = 2.92MRR1878 pKa = 11.84LAEE1881 pKa = 4.84LDD1883 pKa = 3.43NDD1885 pKa = 3.34VWTMEE1890 pKa = 3.81RR1891 pKa = 11.84GYY1893 pKa = 10.92RR1894 pKa = 11.84GATSSAMAMAGLPQAYY1910 pKa = 10.01LPGKK1914 pKa = 10.56SMLAVGFGGYY1924 pKa = 8.88QGEE1927 pKa = 4.42YY1928 pKa = 10.18GMAVGLSGITDD1939 pKa = 3.41NGRR1942 pKa = 11.84WVYY1945 pKa = 10.26KK1946 pKa = 10.62ASASGNTSRR1955 pKa = 11.84DD1956 pKa = 2.75WGFSVGAGIQWW1967 pKa = 3.17
SS1 pKa = 6.03VGSVGAEE8 pKa = 3.35RR9 pKa = 11.84TITNVAAGRR18 pKa = 11.84ISGSSTDD25 pKa = 4.69AINGSQLFATNQAIAAIAASTPVHH49 pKa = 5.88YY50 pKa = 10.42FSVNDD55 pKa = 4.71GGTQQANYY63 pKa = 10.08LNDD66 pKa = 3.61GATAPFAIAIGSNATATGSYY86 pKa = 7.95GTAVGEE92 pKa = 4.24GATAIGDD99 pKa = 3.48WATAYY104 pKa = 10.73GNDD107 pKa = 3.68ANAGTTTATAIGAGTSATATFAFGAGFQATASGVSSTAVGLHH149 pKa = 6.25ANASGEE155 pKa = 4.16SGIAVGTDD163 pKa = 2.9AASTGTYY170 pKa = 10.17AIAIGRR176 pKa = 11.84NASATGSHH184 pKa = 5.36GTAVGEE190 pKa = 4.2GATAIGDD197 pKa = 3.48WATAYY202 pKa = 10.75GNGANASTTTATAIGGGTSATATFAFGAGYY232 pKa = 9.67QATAGGIASTAVGLHH247 pKa = 6.15ANATGEE253 pKa = 4.07SSIAIGTDD261 pKa = 3.03AVADD265 pKa = 3.7SDD267 pKa = 3.91EE268 pKa = 4.14AVAIGRR274 pKa = 11.84GSVATGGRR282 pKa = 11.84AVAIGSGNVANGDD295 pKa = 3.61GAVAIGDD302 pKa = 4.13PNTATGNGAIASGLDD317 pKa = 3.17NTATGNGSVAMGNTNMVGGGGQAVSTPGTAAQGAVGIGYY356 pKa = 9.49QNTVIGQGSVALGSTSSALAAGAVAFGDD384 pKa = 3.51TAVANNAGDD393 pKa = 3.63VALGSGSVTDD403 pKa = 3.76TAVATASTVIDD414 pKa = 3.59GVTYY418 pKa = 10.85NFAGTTPTSTVSVGAVGSEE437 pKa = 3.58RR438 pKa = 11.84TITNVAAGRR447 pKa = 11.84ISDD450 pKa = 3.88TSTDD454 pKa = 4.24AINGSQLYY462 pKa = 8.24ATNQAIEE469 pKa = 4.26AVATTASAGWNLSAQGANATNVAPDD494 pKa = 3.44EE495 pKa = 4.33TVDD498 pKa = 5.09LNNSDD503 pKa = 3.65GNIVVSKK510 pKa = 9.28TATDD514 pKa = 3.82DD515 pKa = 4.84DD516 pKa = 4.4VTFDD520 pKa = 4.12LAADD524 pKa = 3.52ITVDD528 pKa = 3.32SVTAGGTVLNTNGLTIAGGPSVTTTGINAGGLVISNVAPGVAGTDD573 pKa = 3.37AVNVDD578 pKa = 3.45QLTSVSTAGLNFTGNDD594 pKa = 3.08NTAGDD599 pKa = 3.62VHH601 pKa = 7.43RR602 pKa = 11.84DD603 pKa = 3.27LGQTLAIQGGASTAGTYY620 pKa = 10.39SGGNLRR626 pKa = 11.84TVTDD630 pKa = 4.13PVTGAIQLQMADD642 pKa = 3.75SPVFGATTVNAGGTGIISGVTAGVADD668 pKa = 4.0TDD670 pKa = 4.0AVNVSQLNAIGDD682 pKa = 3.75IANAGWNISAQGANATNVAPDD703 pKa = 3.44EE704 pKa = 4.33TVDD707 pKa = 5.09LNNSDD712 pKa = 3.65GNIVVSKK719 pKa = 9.28TATDD723 pKa = 3.82DD724 pKa = 4.84DD725 pKa = 4.4VTFDD729 pKa = 4.12LAADD733 pKa = 3.52ITVDD737 pKa = 3.78SVTAGNSVLNTSGLTVDD754 pKa = 5.19DD755 pKa = 4.84GVNSTSVGSTGLVITGGPSVTTTGIDD781 pKa = 3.07AGSLVISNVAPGVAGTDD798 pKa = 3.37AVNVDD803 pKa = 3.45QLTSVSTAGLNFTGNDD819 pKa = 3.08NTAGDD824 pKa = 3.62VHH826 pKa = 7.43RR827 pKa = 11.84DD828 pKa = 3.27LGQTLAIRR836 pKa = 11.84GGASTAGTYY845 pKa = 10.39SGGNLRR851 pKa = 11.84TVTDD855 pKa = 4.01PATGAIQLQMADD867 pKa = 3.56SPVFGGTTVNAGGTGIISGVTAGVADD893 pKa = 4.0TDD895 pKa = 4.0AVNVSQLNAIGDD907 pKa = 3.74IATAGWNLSAQGANATNVAPDD928 pKa = 3.44EE929 pKa = 4.33TVDD932 pKa = 5.09LNNSDD937 pKa = 3.65GNIVVSKK944 pKa = 10.71NATDD948 pKa = 4.25DD949 pKa = 4.77DD950 pKa = 4.54VTFDD954 pKa = 4.04LADD957 pKa = 5.61DD958 pKa = 3.76ITVDD962 pKa = 3.41SVTAGGTVLNSNGLTIAGGPSVTTTGINAGGLVISNVAPGVAGTDD1007 pKa = 3.37AVNVDD1012 pKa = 3.25QLTAVGNIANAGWNITDD1029 pKa = 3.84GTNSGNIAPNEE1040 pKa = 4.2TLTVAAGSNATVVYY1054 pKa = 10.29DD1055 pKa = 4.38DD1056 pKa = 3.78STGTMTVGVVADD1068 pKa = 4.12PSFNSITVGDD1078 pKa = 3.89TVINDD1083 pKa = 3.41GSISFTSGGPSITNIGIDD1101 pKa = 3.75AGNTTITNVAAAVNNTDD1118 pKa = 3.52AVNLEE1123 pKa = 4.05QVTNLVEE1130 pKa = 4.49GATTRR1135 pKa = 11.84YY1136 pKa = 10.47YY1137 pKa = 11.01SVNDD1141 pKa = 3.38NGVQGGNYY1149 pKa = 9.72NNDD1152 pKa = 3.02GATGINAMAAGVGASSAGAQGVALGNNASVITDD1185 pKa = 3.7GGVALGSGSLSDD1197 pKa = 4.39RR1198 pKa = 11.84VVAPQSGGIIAGTGLIPYY1216 pKa = 7.62NTTDD1220 pKa = 3.11KK1221 pKa = 10.43TLLGAVSVGTDD1232 pKa = 2.96TSYY1235 pKa = 11.36RR1236 pKa = 11.84QITNVADD1243 pKa = 3.7GTEE1246 pKa = 4.21DD1247 pKa = 3.21QDD1249 pKa = 5.01AVTLRR1254 pKa = 11.84QLRR1257 pKa = 11.84NGLSSFAVTPTMYY1270 pKa = 10.49FHH1272 pKa = 7.45ANSTAPDD1279 pKa = 3.48SLAIGAEE1286 pKa = 4.33SVAIGPNTTVNGDD1299 pKa = 3.18NGIGMGNGALVQQTAPGGIAIGQDD1323 pKa = 3.5TIVNFADD1330 pKa = 4.6GVALGTNAVTNGIQAMALGAGATANEE1356 pKa = 4.54PGSVALGAGSVTAVAVATPSTTIDD1380 pKa = 3.1GVTYY1384 pKa = 10.32NYY1386 pKa = 10.9AGTAPTSTVSVGAAGTEE1403 pKa = 3.85RR1404 pKa = 11.84TITHH1408 pKa = 7.08LAAGRR1413 pKa = 11.84ISADD1417 pKa = 2.96STDD1420 pKa = 3.76AVNGSQLFAAHH1431 pKa = 6.1QSIEE1435 pKa = 4.1NLSTRR1440 pKa = 11.84IDD1442 pKa = 3.46ASATHH1447 pKa = 6.45YY1448 pKa = 11.15YY1449 pKa = 10.24SVNDD1453 pKa = 3.53NGVVGGNYY1461 pKa = 10.14NNDD1464 pKa = 3.12GATGINSLATGVDD1477 pKa = 3.7SAASGDD1483 pKa = 3.49ASVAIGFGATAADD1496 pKa = 3.99ANSVALGSNATTAAAVGTASIVIGGTTYY1524 pKa = 11.23NFAGTAPVGTVSVGDD1539 pKa = 3.55VGAEE1543 pKa = 3.55RR1544 pKa = 11.84TITNVAAGRR1553 pKa = 11.84IGADD1557 pKa = 2.83STDD1560 pKa = 3.41AVNGSQLHH1568 pKa = 5.69ATNQAIANLSNTVNASTAHH1587 pKa = 5.95YY1588 pKa = 10.49YY1589 pKa = 10.36SVNDD1593 pKa = 3.9GGVQGGNYY1601 pKa = 9.87NNDD1604 pKa = 3.26GASGVNAVASGVNAAASGQGSVAVGANSNAAGAGSVAVGSGSQSTVAGGVAIGADD1659 pKa = 3.88SVSSTAAGVAGYY1671 pKa = 10.28VPAGATAGQAAAIAATTSTSGAVSVGDD1698 pKa = 3.99ASSGLYY1704 pKa = 9.79RR1705 pKa = 11.84QITGVAAGTAASDD1718 pKa = 3.81AVNVAQLQAVQNQLNATATHH1738 pKa = 6.57YY1739 pKa = 11.18YY1740 pKa = 9.73SVNDD1744 pKa = 3.92GGVQGGNYY1752 pKa = 9.8NNDD1755 pKa = 3.03GATGVNSMAGGVNSTASGSNSVALGANSNASAANSAALGSGSQATHH1801 pKa = 5.76VNSVAIGNGSATTVGAQTGYY1821 pKa = 9.17NAAYY1825 pKa = 9.9VGSSNSTGEE1834 pKa = 3.89VNMGGRR1840 pKa = 11.84TITGVAPGIAGTDD1853 pKa = 3.24AVNVNQLNAGVNHH1866 pKa = 7.21AVNQANQYY1874 pKa = 9.04TDD1876 pKa = 2.92MRR1878 pKa = 11.84LAEE1881 pKa = 4.84LDD1883 pKa = 3.43NDD1885 pKa = 3.34VWTMEE1890 pKa = 3.81RR1891 pKa = 11.84GYY1893 pKa = 10.92RR1894 pKa = 11.84GATSSAMAMAGLPQAYY1910 pKa = 10.01LPGKK1914 pKa = 10.56SMLAVGFGGYY1924 pKa = 8.88QGEE1927 pKa = 4.42YY1928 pKa = 10.18GMAVGLSGITDD1939 pKa = 3.41NGRR1942 pKa = 11.84WVYY1945 pKa = 10.26KK1946 pKa = 10.62ASASGNTSRR1955 pKa = 11.84DD1956 pKa = 2.75WGFSVGAGIQWW1967 pKa = 3.17
Molecular weight: 188.29 kDa
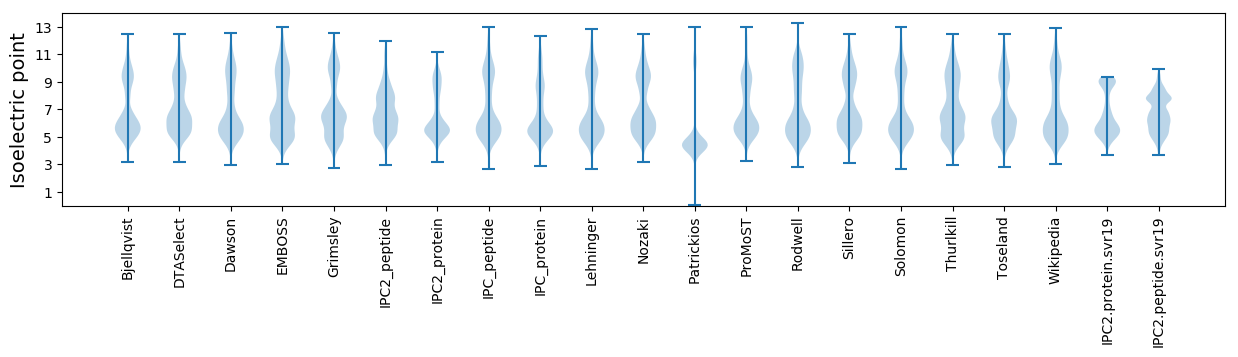
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A286DBA4|A0A286DBA4_9GAMM Chemotaxis protein MotA OS=Pseudoxanthomonas wuyuanensis OX=1073196 GN=SAMN06296416_10886 PE=3 SV=1
MM1 pKa = 7.37LNSVAVGRR9 pKa = 11.84RR10 pKa = 11.84LALRR14 pKa = 11.84AAAYY18 pKa = 9.8QLLAVGLVAALFLLRR33 pKa = 11.84GLPQALAAALGGLAMLAGGLVAARR57 pKa = 11.84LALGGGVIPAGAAMLRR73 pKa = 11.84LLAGVVLKK81 pKa = 9.97WAVVFAVLLVGLGAWKK97 pKa = 10.32LPPLPLMAGLLTGLAAQVLAAARR120 pKa = 11.84RR121 pKa = 11.84QQ122 pKa = 3.35
MM1 pKa = 7.37LNSVAVGRR9 pKa = 11.84RR10 pKa = 11.84LALRR14 pKa = 11.84AAAYY18 pKa = 9.8QLLAVGLVAALFLLRR33 pKa = 11.84GLPQALAAALGGLAMLAGGLVAARR57 pKa = 11.84LALGGGVIPAGAAMLRR73 pKa = 11.84LLAGVVLKK81 pKa = 9.97WAVVFAVLLVGLGAWKK97 pKa = 10.32LPPLPLMAGLLTGLAAQVLAAARR120 pKa = 11.84RR121 pKa = 11.84QQ122 pKa = 3.35
Molecular weight: 12.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1357910 |
29 |
4271 |
340.4 |
37.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.97 ± 0.061 | 0.804 ± 0.013 |
5.756 ± 0.03 | 5.315 ± 0.035 |
3.42 ± 0.02 | 8.631 ± 0.053 |
2.115 ± 0.022 | 4.246 ± 0.028 |
2.669 ± 0.032 | 10.79 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.117 ± 0.017 | 2.629 ± 0.035 |
5.381 ± 0.031 | 4.032 ± 0.024 |
7.609 ± 0.047 | 5.483 ± 0.033 |
4.86 ± 0.05 | 7.092 ± 0.031 |
1.587 ± 0.019 | 2.493 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
