
Gluconobacter oxydans (strain 621H) (Gluconobacter suboxydans)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Gluconobacter; Gluconobacter oxydans
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
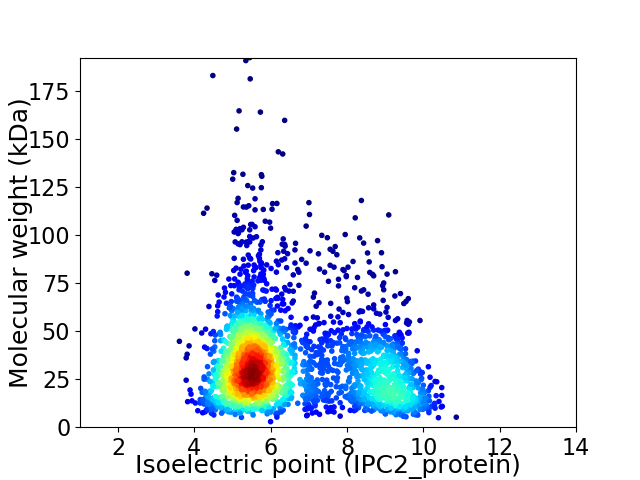
Virtual 2D-PAGE plot for 2626 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
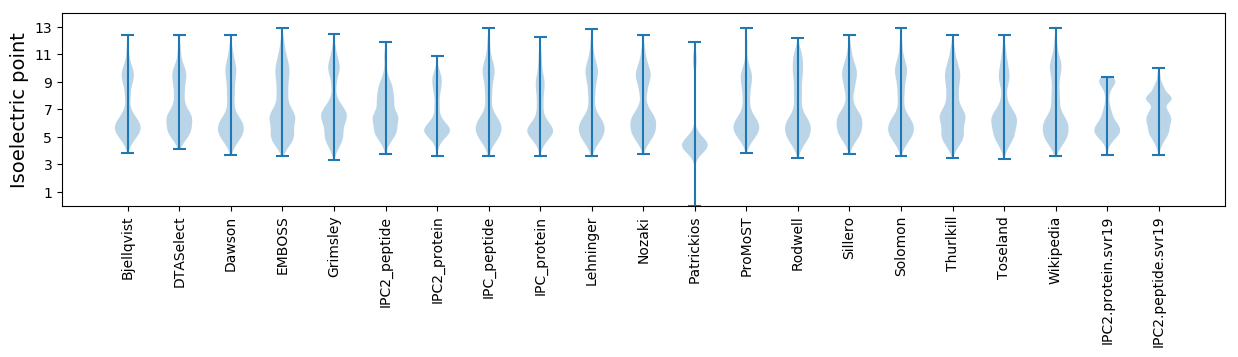
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q5FS93|Q5FS93_GLUOX 6 7-dimethyl-8-ribityllumazine synthase OS=Gluconobacter oxydans (strain 621H) OX=290633 GN=ribH PE=3 SV=1
MM1 pKa = 7.47SSIINAVHH9 pKa = 5.79TAVSGLNAQSHH20 pKa = 6.39AFTDD24 pKa = 4.34LSNNIANSQTTGYY37 pKa = 9.63KK38 pKa = 10.4ASTTSFEE45 pKa = 5.37DD46 pKa = 3.9YY47 pKa = 9.44VTNNEE52 pKa = 4.17LANDD56 pKa = 4.45GEE58 pKa = 4.54SLSDD62 pKa = 3.48SVAATTRR69 pKa = 11.84QHH71 pKa = 6.48TDD73 pKa = 2.99NQCMVTTSTNSTALAISGNGFFNVVQATGQSNDD106 pKa = 3.29STPVLGTQQYY116 pKa = 6.44YY117 pKa = 8.86TRR119 pKa = 11.84NGDD122 pKa = 3.59FSQDD126 pKa = 3.07KK127 pKa = 10.73NGYY130 pKa = 8.72LVNTSGYY137 pKa = 8.81YY138 pKa = 10.12LEE140 pKa = 5.47GYY142 pKa = 9.83NVDD145 pKa = 3.64ATSGTLGTSLSAIQVPSTIAFRR167 pKa = 11.84PTEE170 pKa = 4.27STKK173 pKa = 10.15LTATGTLDD181 pKa = 3.27SSSSKK186 pKa = 9.35TVDD189 pKa = 3.1MSTTAYY195 pKa = 10.42DD196 pKa = 3.56STGAASTVDD205 pKa = 3.78LSWTKK210 pKa = 10.97LSDD213 pKa = 3.44TTWAVKK219 pKa = 10.1DD220 pKa = 3.96VNDD223 pKa = 3.8TTGTSNSATVTFDD236 pKa = 2.8SSGAVSSVTSQDD248 pKa = 3.06STGASSTSSSSFNYY262 pKa = 9.31TGSPQDD268 pKa = 3.39MTVSLSGVTLGSANTVSMTSDD289 pKa = 2.89SVTTGTFSGISMEE302 pKa = 4.27TDD304 pKa = 3.13GSVMATFDD312 pKa = 3.2NGYY315 pKa = 8.5SQLVAKK321 pKa = 10.53IPLTTFADD329 pKa = 4.12PNSLSAQNGQAYY341 pKa = 6.97TATSQSGSPQINAVNTNGAGTLTTGSIEE369 pKa = 4.31EE370 pKa = 4.47STTDD374 pKa = 3.46LTGDD378 pKa = 3.96LSKK381 pKa = 11.22LIVAQQAYY389 pKa = 8.94GANTKK394 pKa = 9.73VVSTADD400 pKa = 3.32QLLQTTLAMIQQ411 pKa = 3.18
MM1 pKa = 7.47SSIINAVHH9 pKa = 5.79TAVSGLNAQSHH20 pKa = 6.39AFTDD24 pKa = 4.34LSNNIANSQTTGYY37 pKa = 9.63KK38 pKa = 10.4ASTTSFEE45 pKa = 5.37DD46 pKa = 3.9YY47 pKa = 9.44VTNNEE52 pKa = 4.17LANDD56 pKa = 4.45GEE58 pKa = 4.54SLSDD62 pKa = 3.48SVAATTRR69 pKa = 11.84QHH71 pKa = 6.48TDD73 pKa = 2.99NQCMVTTSTNSTALAISGNGFFNVVQATGQSNDD106 pKa = 3.29STPVLGTQQYY116 pKa = 6.44YY117 pKa = 8.86TRR119 pKa = 11.84NGDD122 pKa = 3.59FSQDD126 pKa = 3.07KK127 pKa = 10.73NGYY130 pKa = 8.72LVNTSGYY137 pKa = 8.81YY138 pKa = 10.12LEE140 pKa = 5.47GYY142 pKa = 9.83NVDD145 pKa = 3.64ATSGTLGTSLSAIQVPSTIAFRR167 pKa = 11.84PTEE170 pKa = 4.27STKK173 pKa = 10.15LTATGTLDD181 pKa = 3.27SSSSKK186 pKa = 9.35TVDD189 pKa = 3.1MSTTAYY195 pKa = 10.42DD196 pKa = 3.56STGAASTVDD205 pKa = 3.78LSWTKK210 pKa = 10.97LSDD213 pKa = 3.44TTWAVKK219 pKa = 10.1DD220 pKa = 3.96VNDD223 pKa = 3.8TTGTSNSATVTFDD236 pKa = 2.8SSGAVSSVTSQDD248 pKa = 3.06STGASSTSSSSFNYY262 pKa = 9.31TGSPQDD268 pKa = 3.39MTVSLSGVTLGSANTVSMTSDD289 pKa = 2.89SVTTGTFSGISMEE302 pKa = 4.27TDD304 pKa = 3.13GSVMATFDD312 pKa = 3.2NGYY315 pKa = 8.5SQLVAKK321 pKa = 10.53IPLTTFADD329 pKa = 4.12PNSLSAQNGQAYY341 pKa = 6.97TATSQSGSPQINAVNTNGAGTLTTGSIEE369 pKa = 4.31EE370 pKa = 4.47STTDD374 pKa = 3.46LTGDD378 pKa = 3.96LSKK381 pKa = 11.22LIVAQQAYY389 pKa = 8.94GANTKK394 pKa = 9.73VVSTADD400 pKa = 3.32QLLQTTLAMIQQ411 pKa = 3.18
Molecular weight: 42.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q5FTT2|Q5FTT2_GLUOX Phosphogluconate dehydratase OS=Gluconobacter oxydans (strain 621H) OX=290633 GN=edd PE=3 SV=1
MM1 pKa = 7.54GFFRR5 pKa = 11.84TFRR8 pKa = 11.84DD9 pKa = 4.17GYY11 pKa = 10.55LVLRR15 pKa = 11.84FRR17 pKa = 11.84NWEE20 pKa = 3.78IQKK23 pKa = 9.53VVHH26 pKa = 5.44FHH28 pKa = 6.56GYY30 pKa = 9.86SPVSRR35 pKa = 11.84LRR37 pKa = 11.84SLDD40 pKa = 3.54LPRR43 pKa = 11.84SLVVRR48 pKa = 11.84RR49 pKa = 11.84VVALDD54 pKa = 3.21NRR56 pKa = 11.84VEE58 pKa = 4.11IEE60 pKa = 3.68VGLRR64 pKa = 11.84KK65 pKa = 9.87RR66 pKa = 11.84SAVCPDD72 pKa = 4.08CQCPTQRR79 pKa = 11.84IHH81 pKa = 7.38SFYY84 pKa = 10.36QRR86 pKa = 11.84KK87 pKa = 8.54LADD90 pKa = 4.65LPWQSRR96 pKa = 11.84PATLIVSVPRR106 pKa = 11.84FFCQTMLCPRR116 pKa = 11.84RR117 pKa = 11.84TFVMPLEE124 pKa = 5.08GITVRR129 pKa = 11.84HH130 pKa = 5.66GRR132 pKa = 11.84QTTRR136 pKa = 11.84LADD139 pKa = 3.18LHH141 pKa = 6.76YY142 pKa = 10.7YY143 pKa = 8.98IAHH146 pKa = 6.38TLGGSAGARR155 pKa = 11.84MTVRR159 pKa = 11.84LCCPISADD167 pKa = 3.52TLVRR171 pKa = 11.84RR172 pKa = 11.84LLSRR176 pKa = 11.84VQNTTKK182 pKa = 9.73GTALTRR188 pKa = 11.84VVGVDD193 pKa = 2.97DD194 pKa = 3.8WAWRR198 pKa = 11.84RR199 pKa = 11.84GHH201 pKa = 6.76HH202 pKa = 6.33YY203 pKa = 9.33GTIVVDD209 pKa = 3.89LEE211 pKa = 4.35KK212 pKa = 11.17NDD214 pKa = 4.57VIDD217 pKa = 5.12LLPDD221 pKa = 3.71RR222 pKa = 11.84DD223 pKa = 4.22ADD225 pKa = 3.92TLARR229 pKa = 11.84WLQVHH234 pKa = 6.72PGIEE238 pKa = 3.92IIARR242 pKa = 11.84DD243 pKa = 3.57RR244 pKa = 11.84AGTYY248 pKa = 10.7ADD250 pKa = 3.52GCRR253 pKa = 11.84RR254 pKa = 11.84GAPSALQVTDD264 pKa = 3.09RR265 pKa = 11.84WHH267 pKa = 6.23LLKK270 pKa = 10.8NLSDD274 pKa = 3.92AFVSILDD281 pKa = 3.7RR282 pKa = 11.84FTTTVRR288 pKa = 11.84TLTRR292 pKa = 11.84QMNVSTVVSKK302 pKa = 10.78ISEE305 pKa = 4.51TIRR308 pKa = 11.84AHH310 pKa = 6.13PEE312 pKa = 3.15EE313 pKa = 4.41AQAVVRR319 pKa = 11.84SASGEE324 pKa = 3.83RR325 pKa = 11.84RR326 pKa = 11.84DD327 pKa = 3.89ILFKK331 pKa = 10.74EE332 pKa = 4.05ALALKK337 pKa = 10.2AAGHH341 pKa = 6.64SIQAIATTIGVEE353 pKa = 3.96RR354 pKa = 11.84KK355 pKa = 7.01TVRR358 pKa = 11.84RR359 pKa = 11.84WFQRR363 pKa = 11.84GHH365 pKa = 5.9APPWKK370 pKa = 8.31RR371 pKa = 11.84TVPSRR376 pKa = 11.84SILVPYY382 pKa = 8.12TAHH385 pKa = 7.4LEE387 pKa = 4.29KK388 pKa = 10.3RR389 pKa = 11.84WSEE392 pKa = 3.84GCRR395 pKa = 11.84NSRR398 pKa = 11.84QLWRR402 pKa = 11.84EE403 pKa = 3.65LLEE406 pKa = 4.0QGFAGQYY413 pKa = 6.71GTVWKK418 pKa = 9.66WVATRR423 pKa = 11.84QRR425 pKa = 11.84CSAKK429 pKa = 9.6PVPSAHH435 pKa = 5.24VVAPRR440 pKa = 11.84GRR442 pKa = 11.84RR443 pKa = 11.84LARR446 pKa = 11.84LLLAEE451 pKa = 5.31DD452 pKa = 4.45PLSAGQEE459 pKa = 4.27GLLVPALLEE468 pKa = 4.45AEE470 pKa = 4.1PHH472 pKa = 6.28LAVTLCWLRR481 pKa = 11.84KK482 pKa = 8.18MQTLLCKK489 pKa = 10.5RR490 pKa = 11.84GDD492 pKa = 3.52TCTGGDD498 pKa = 3.65LKK500 pKa = 11.36ALLEE504 pKa = 4.29EE505 pKa = 5.0GKK507 pKa = 7.51TTLLSRR513 pKa = 11.84LIPALEE519 pKa = 4.11RR520 pKa = 11.84DD521 pKa = 3.44AAAIEE526 pKa = 4.3ASLVTPWTTSPVEE539 pKa = 4.1GQISRR544 pKa = 11.84LKK546 pKa = 9.4MIKK549 pKa = 8.82RR550 pKa = 11.84TMFGRR555 pKa = 11.84AGFEE559 pKa = 3.95LLRR562 pKa = 11.84ARR564 pKa = 11.84VLQPAA569 pKa = 3.85
MM1 pKa = 7.54GFFRR5 pKa = 11.84TFRR8 pKa = 11.84DD9 pKa = 4.17GYY11 pKa = 10.55LVLRR15 pKa = 11.84FRR17 pKa = 11.84NWEE20 pKa = 3.78IQKK23 pKa = 9.53VVHH26 pKa = 5.44FHH28 pKa = 6.56GYY30 pKa = 9.86SPVSRR35 pKa = 11.84LRR37 pKa = 11.84SLDD40 pKa = 3.54LPRR43 pKa = 11.84SLVVRR48 pKa = 11.84RR49 pKa = 11.84VVALDD54 pKa = 3.21NRR56 pKa = 11.84VEE58 pKa = 4.11IEE60 pKa = 3.68VGLRR64 pKa = 11.84KK65 pKa = 9.87RR66 pKa = 11.84SAVCPDD72 pKa = 4.08CQCPTQRR79 pKa = 11.84IHH81 pKa = 7.38SFYY84 pKa = 10.36QRR86 pKa = 11.84KK87 pKa = 8.54LADD90 pKa = 4.65LPWQSRR96 pKa = 11.84PATLIVSVPRR106 pKa = 11.84FFCQTMLCPRR116 pKa = 11.84RR117 pKa = 11.84TFVMPLEE124 pKa = 5.08GITVRR129 pKa = 11.84HH130 pKa = 5.66GRR132 pKa = 11.84QTTRR136 pKa = 11.84LADD139 pKa = 3.18LHH141 pKa = 6.76YY142 pKa = 10.7YY143 pKa = 8.98IAHH146 pKa = 6.38TLGGSAGARR155 pKa = 11.84MTVRR159 pKa = 11.84LCCPISADD167 pKa = 3.52TLVRR171 pKa = 11.84RR172 pKa = 11.84LLSRR176 pKa = 11.84VQNTTKK182 pKa = 9.73GTALTRR188 pKa = 11.84VVGVDD193 pKa = 2.97DD194 pKa = 3.8WAWRR198 pKa = 11.84RR199 pKa = 11.84GHH201 pKa = 6.76HH202 pKa = 6.33YY203 pKa = 9.33GTIVVDD209 pKa = 3.89LEE211 pKa = 4.35KK212 pKa = 11.17NDD214 pKa = 4.57VIDD217 pKa = 5.12LLPDD221 pKa = 3.71RR222 pKa = 11.84DD223 pKa = 4.22ADD225 pKa = 3.92TLARR229 pKa = 11.84WLQVHH234 pKa = 6.72PGIEE238 pKa = 3.92IIARR242 pKa = 11.84DD243 pKa = 3.57RR244 pKa = 11.84AGTYY248 pKa = 10.7ADD250 pKa = 3.52GCRR253 pKa = 11.84RR254 pKa = 11.84GAPSALQVTDD264 pKa = 3.09RR265 pKa = 11.84WHH267 pKa = 6.23LLKK270 pKa = 10.8NLSDD274 pKa = 3.92AFVSILDD281 pKa = 3.7RR282 pKa = 11.84FTTTVRR288 pKa = 11.84TLTRR292 pKa = 11.84QMNVSTVVSKK302 pKa = 10.78ISEE305 pKa = 4.51TIRR308 pKa = 11.84AHH310 pKa = 6.13PEE312 pKa = 3.15EE313 pKa = 4.41AQAVVRR319 pKa = 11.84SASGEE324 pKa = 3.83RR325 pKa = 11.84RR326 pKa = 11.84DD327 pKa = 3.89ILFKK331 pKa = 10.74EE332 pKa = 4.05ALALKK337 pKa = 10.2AAGHH341 pKa = 6.64SIQAIATTIGVEE353 pKa = 3.96RR354 pKa = 11.84KK355 pKa = 7.01TVRR358 pKa = 11.84RR359 pKa = 11.84WFQRR363 pKa = 11.84GHH365 pKa = 5.9APPWKK370 pKa = 8.31RR371 pKa = 11.84TVPSRR376 pKa = 11.84SILVPYY382 pKa = 8.12TAHH385 pKa = 7.4LEE387 pKa = 4.29KK388 pKa = 10.3RR389 pKa = 11.84WSEE392 pKa = 3.84GCRR395 pKa = 11.84NSRR398 pKa = 11.84QLWRR402 pKa = 11.84EE403 pKa = 3.65LLEE406 pKa = 4.0QGFAGQYY413 pKa = 6.71GTVWKK418 pKa = 9.66WVATRR423 pKa = 11.84QRR425 pKa = 11.84CSAKK429 pKa = 9.6PVPSAHH435 pKa = 5.24VVAPRR440 pKa = 11.84GRR442 pKa = 11.84RR443 pKa = 11.84LARR446 pKa = 11.84LLLAEE451 pKa = 5.31DD452 pKa = 4.45PLSAGQEE459 pKa = 4.27GLLVPALLEE468 pKa = 4.45AEE470 pKa = 4.1PHH472 pKa = 6.28LAVTLCWLRR481 pKa = 11.84KK482 pKa = 8.18MQTLLCKK489 pKa = 10.5RR490 pKa = 11.84GDD492 pKa = 3.52TCTGGDD498 pKa = 3.65LKK500 pKa = 11.36ALLEE504 pKa = 4.29EE505 pKa = 5.0GKK507 pKa = 7.51TTLLSRR513 pKa = 11.84LIPALEE519 pKa = 4.11RR520 pKa = 11.84DD521 pKa = 3.44AAAIEE526 pKa = 4.3ASLVTPWTTSPVEE539 pKa = 4.1GQISRR544 pKa = 11.84LKK546 pKa = 9.4MIKK549 pKa = 8.82RR550 pKa = 11.84TMFGRR555 pKa = 11.84AGFEE559 pKa = 3.95LLRR562 pKa = 11.84ARR564 pKa = 11.84VLQPAA569 pKa = 3.85
Molecular weight: 64.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
859884 |
26 |
1750 |
327.5 |
35.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.264 ± 0.062 | 1.029 ± 0.017 |
5.596 ± 0.039 | 5.356 ± 0.049 |
3.483 ± 0.029 | 8.349 ± 0.05 |
2.418 ± 0.023 | 4.93 ± 0.036 |
3.006 ± 0.037 | 10.317 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.538 ± 0.022 | 2.729 ± 0.034 |
5.602 ± 0.037 | 3.563 ± 0.027 |
7.213 ± 0.048 | 6.1 ± 0.043 |
5.897 ± 0.047 | 7.045 ± 0.042 |
1.381 ± 0.018 | 2.183 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
