
bacterium HR31
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 7.36
Get precalculated fractions of proteins
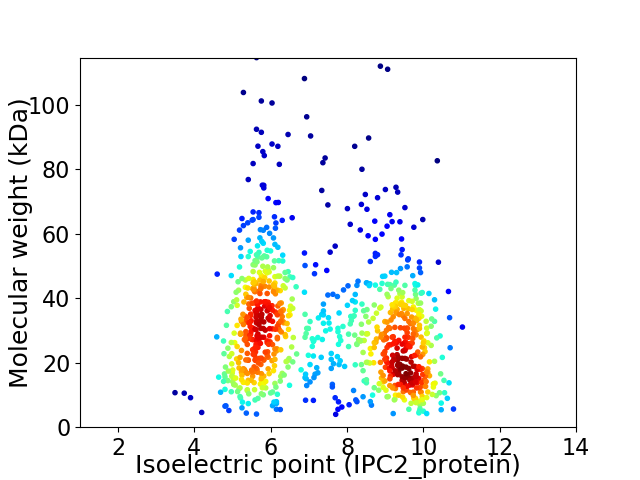
Virtual 2D-PAGE plot for 927 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
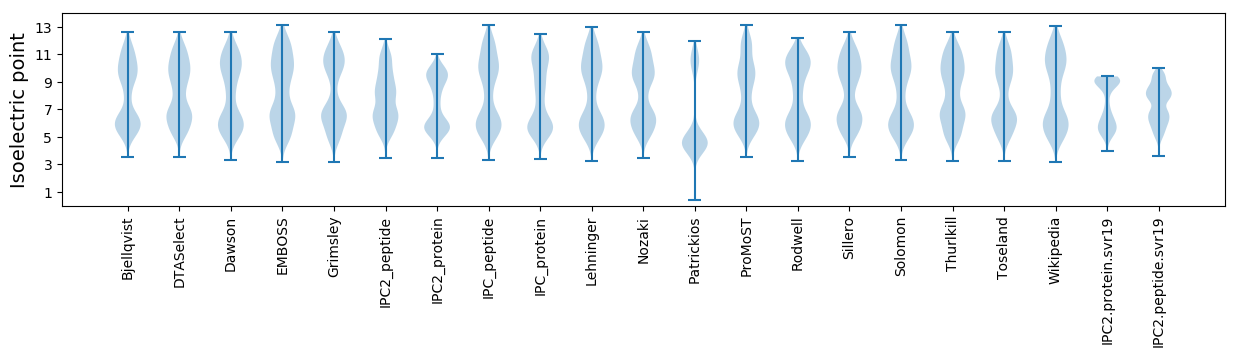
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5ZR23|A0A2H5ZR23_9BACT Cell division ATP-binding protein FtsE OS=bacterium HR31 OX=2035426 GN=ftsE PE=3 SV=1
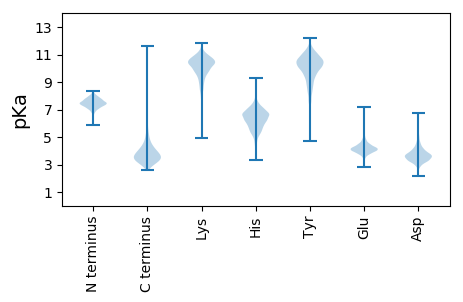
MM1 pKa = 7.3TVPFNRR7 pKa = 11.84PCPQCGGEE15 pKa = 4.15MEE17 pKa = 5.18LLASDD22 pKa = 3.79EE23 pKa = 4.34LGAVVVYY30 pKa = 9.61EE31 pKa = 4.25CAEE34 pKa = 4.01CGYY37 pKa = 8.89QAKK40 pKa = 10.02EE41 pKa = 3.73RR42 pKa = 11.84VEE44 pKa = 4.34EE45 pKa = 3.99VLEE48 pKa = 4.05PAEE51 pKa = 4.15EE52 pKa = 4.51DD53 pKa = 3.63EE54 pKa = 5.09VALQPEE60 pKa = 4.52EE61 pKa = 4.24EE62 pKa = 4.36EE63 pKa = 4.2EE64 pKa = 4.35EE65 pKa = 4.19EE66 pKa = 4.3FPEE69 pKa = 4.27EE70 pKa = 4.14EE71 pKa = 3.88EE72 pKa = 4.8DD73 pKa = 5.05EE74 pKa = 4.29EE75 pKa = 5.59FEE77 pKa = 4.77DD78 pKa = 5.36SLWEE82 pKa = 4.29DD83 pKa = 3.6EE84 pKa = 4.99EE85 pKa = 4.56EE86 pKa = 5.83EE87 pKa = 3.92EE88 pKa = 5.22DD89 pKa = 3.28AWRR92 pKa = 4.28
MM1 pKa = 7.3TVPFNRR7 pKa = 11.84PCPQCGGEE15 pKa = 4.15MEE17 pKa = 5.18LLASDD22 pKa = 3.79EE23 pKa = 4.34LGAVVVYY30 pKa = 9.61EE31 pKa = 4.25CAEE34 pKa = 4.01CGYY37 pKa = 8.89QAKK40 pKa = 10.02EE41 pKa = 3.73RR42 pKa = 11.84VEE44 pKa = 4.34EE45 pKa = 3.99VLEE48 pKa = 4.05PAEE51 pKa = 4.15EE52 pKa = 4.51DD53 pKa = 3.63EE54 pKa = 5.09VALQPEE60 pKa = 4.52EE61 pKa = 4.24EE62 pKa = 4.36EE63 pKa = 4.2EE64 pKa = 4.35EE65 pKa = 4.19EE66 pKa = 4.3FPEE69 pKa = 4.27EE70 pKa = 4.14EE71 pKa = 3.88EE72 pKa = 4.8DD73 pKa = 5.05EE74 pKa = 4.29EE75 pKa = 5.59FEE77 pKa = 4.77DD78 pKa = 5.36SLWEE82 pKa = 4.29DD83 pKa = 3.6EE84 pKa = 4.99EE85 pKa = 4.56EE86 pKa = 5.83EE87 pKa = 3.92EE88 pKa = 5.22DD89 pKa = 3.28AWRR92 pKa = 4.28
Molecular weight: 10.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5ZSW6|A0A2H5ZSW6_9BACT Cupin_2 domain-containing protein OS=bacterium HR31 OX=2035426 GN=HRbin31_00832 PE=4 SV=1
MM1 pKa = 7.43GVTKK5 pKa = 10.02PKK7 pKa = 9.99RR8 pKa = 11.84SSRR11 pKa = 11.84YY12 pKa = 8.58GSAPVARR19 pKa = 11.84MPHH22 pKa = 5.58RR23 pKa = 11.84TPPAAPTPALRR34 pKa = 11.84AASRR38 pKa = 11.84VGAPARR44 pKa = 11.84RR45 pKa = 11.84SSASAVAALASPTSRR60 pKa = 11.84PAYY63 pKa = 9.24ARR65 pKa = 11.84SVRR68 pKa = 11.84RR69 pKa = 11.84WIRR72 pKa = 11.84RR73 pKa = 11.84ALNTPTAAPAYY84 pKa = 7.72ATATRR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84SWVPSVPARR100 pKa = 11.84TASAPAAKK108 pKa = 9.81SASRR112 pKa = 11.84GFTSTASRR120 pKa = 11.84MATAAVGTSSGWIVAATRR138 pKa = 11.84RR139 pKa = 11.84VPAAPSHH146 pKa = 5.31TRR148 pKa = 11.84SPVSSRR154 pKa = 11.84SLGLLHH160 pKa = 6.57TSGWKK165 pKa = 6.88WARR168 pKa = 11.84YY169 pKa = 8.96APRR172 pKa = 11.84TATRR176 pKa = 11.84AAASTRR182 pKa = 11.84VPTSLLHH189 pKa = 4.84GHH191 pKa = 6.48GPVASPRR198 pKa = 11.84ACSLRR203 pKa = 11.84ICSGVTTSPRR213 pKa = 11.84RR214 pKa = 11.84TTTSSFRR221 pKa = 11.84TRR223 pKa = 11.84TRR225 pKa = 11.84TGSTPARR232 pKa = 11.84AASRR236 pKa = 11.84ARR238 pKa = 11.84RR239 pKa = 11.84VCSSSTTRR247 pKa = 11.84LGRR250 pKa = 11.84RR251 pKa = 11.84ARR253 pKa = 11.84SATHH257 pKa = 7.02LLNARR262 pKa = 11.84AGPLTPFSRR271 pKa = 11.84RR272 pKa = 11.84IGSSCWRR279 pKa = 11.84ALDD282 pKa = 3.6RR283 pKa = 11.84SSGLRR288 pKa = 11.84GRR290 pKa = 11.84AIKK293 pKa = 10.4RR294 pKa = 3.39
MM1 pKa = 7.43GVTKK5 pKa = 10.02PKK7 pKa = 9.99RR8 pKa = 11.84SSRR11 pKa = 11.84YY12 pKa = 8.58GSAPVARR19 pKa = 11.84MPHH22 pKa = 5.58RR23 pKa = 11.84TPPAAPTPALRR34 pKa = 11.84AASRR38 pKa = 11.84VGAPARR44 pKa = 11.84RR45 pKa = 11.84SSASAVAALASPTSRR60 pKa = 11.84PAYY63 pKa = 9.24ARR65 pKa = 11.84SVRR68 pKa = 11.84RR69 pKa = 11.84WIRR72 pKa = 11.84RR73 pKa = 11.84ALNTPTAAPAYY84 pKa = 7.72ATATRR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84SWVPSVPARR100 pKa = 11.84TASAPAAKK108 pKa = 9.81SASRR112 pKa = 11.84GFTSTASRR120 pKa = 11.84MATAAVGTSSGWIVAATRR138 pKa = 11.84RR139 pKa = 11.84VPAAPSHH146 pKa = 5.31TRR148 pKa = 11.84SPVSSRR154 pKa = 11.84SLGLLHH160 pKa = 6.57TSGWKK165 pKa = 6.88WARR168 pKa = 11.84YY169 pKa = 8.96APRR172 pKa = 11.84TATRR176 pKa = 11.84AAASTRR182 pKa = 11.84VPTSLLHH189 pKa = 4.84GHH191 pKa = 6.48GPVASPRR198 pKa = 11.84ACSLRR203 pKa = 11.84ICSGVTTSPRR213 pKa = 11.84RR214 pKa = 11.84TTTSSFRR221 pKa = 11.84TRR223 pKa = 11.84TRR225 pKa = 11.84TGSTPARR232 pKa = 11.84AASRR236 pKa = 11.84ARR238 pKa = 11.84RR239 pKa = 11.84VCSSSTTRR247 pKa = 11.84LGRR250 pKa = 11.84RR251 pKa = 11.84ARR253 pKa = 11.84SATHH257 pKa = 7.02LLNARR262 pKa = 11.84AGPLTPFSRR271 pKa = 11.84RR272 pKa = 11.84IGSSCWRR279 pKa = 11.84ALDD282 pKa = 3.6RR283 pKa = 11.84SSGLRR288 pKa = 11.84GRR290 pKa = 11.84AIKK293 pKa = 10.4RR294 pKa = 3.39
Molecular weight: 31.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
267116 |
34 |
1041 |
288.2 |
31.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.102 ± 0.097 | 0.891 ± 0.026 |
4.513 ± 0.055 | 6.808 ± 0.093 |
3.038 ± 0.052 | 9.036 ± 0.091 |
2.21 ± 0.062 | 2.72 ± 0.062 |
1.744 ± 0.046 | 11.203 ± 0.114 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.647 ± 0.036 | 1.507 ± 0.033 |
6.152 ± 0.083 | 3.321 ± 0.072 |
10.307 ± 0.099 | 4.039 ± 0.054 |
4.394 ± 0.056 | 10.591 ± 0.081 |
1.471 ± 0.04 | 2.307 ± 0.046 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
