
Fructobacillus durionis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Fructobacillus
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
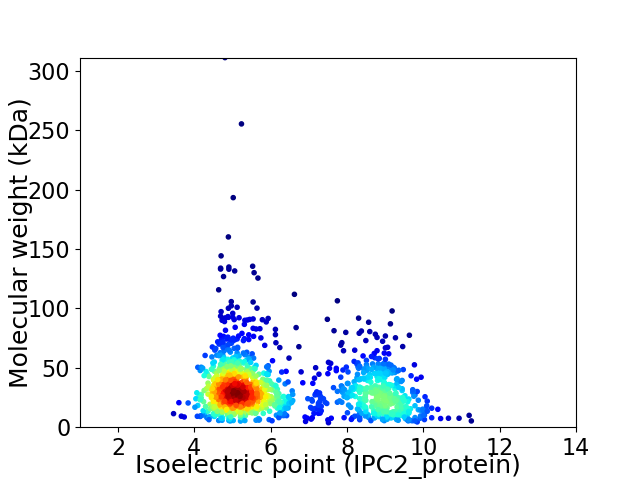
Virtual 2D-PAGE plot for 1250 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
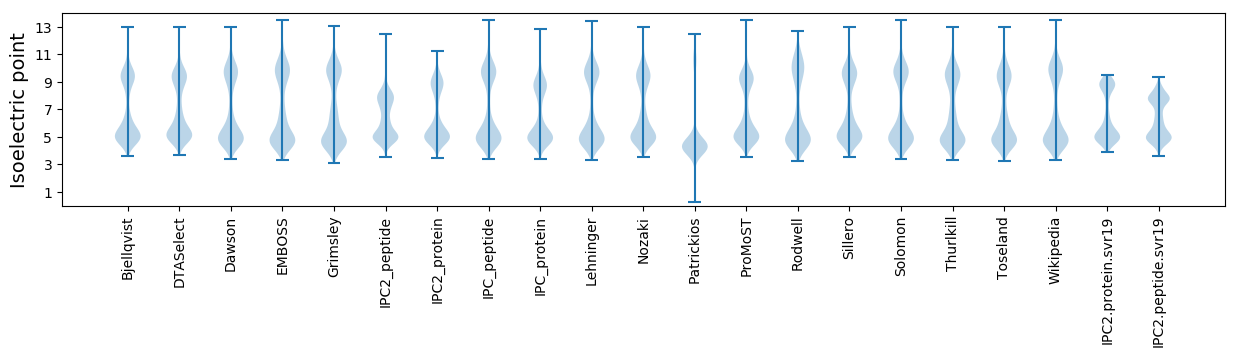
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I1GX56|A0A1I1GX56_9LACO PTS_EIIC domain-containing protein OS=Fructobacillus durionis OX=283737 GN=SAMN05660453_1140 PE=4 SV=1
MM1 pKa = 7.71EE2 pKa = 4.86NQTMKK7 pKa = 10.69QLSKK11 pKa = 11.11RR12 pKa = 11.84LMVTAGASTLAATGAVLGATVVGNAQTASANDD44 pKa = 3.4VTEE47 pKa = 4.39PTSQTDD53 pKa = 4.0AQNQWKK59 pKa = 9.94ANSVDD64 pKa = 3.52HH65 pKa = 6.59VKK67 pKa = 10.36QAVKK71 pKa = 10.24KK72 pKa = 10.53QSAKK76 pKa = 10.42DD77 pKa = 3.51LKK79 pKa = 10.69DD80 pKa = 3.82YY81 pKa = 10.37QVQWGDD87 pKa = 3.52TLSAIAEE94 pKa = 4.35AFGTTADD101 pKa = 4.23AAGQQLGLSDD111 pKa = 4.5HH112 pKa = 6.6GLLVAGQTMGKK123 pKa = 8.51QEE125 pKa = 4.88EE126 pKa = 4.6IIQGLKK132 pKa = 7.65TAGYY136 pKa = 9.43LADD139 pKa = 4.18GQQTQQADD147 pKa = 3.79LGKK150 pKa = 10.54GDD152 pKa = 3.71QNLIMVQQGTPTLDD166 pKa = 3.45TNNGQQTTEE175 pKa = 3.7NTTVNNQTNSQSGSQAAYY193 pKa = 9.3EE194 pKa = 4.2QASQAAAEE202 pKa = 4.28SQAISQSNSMAAATAATNQYY222 pKa = 8.85SQSAASQANNDD233 pKa = 3.46ASQASLTADD242 pKa = 3.4SEE244 pKa = 4.96SQVTPAASDD253 pKa = 3.42ATSTSDD259 pKa = 4.21DD260 pKa = 3.67AAATTPASDD269 pKa = 3.55SSASNSDD276 pKa = 3.14SDD278 pKa = 4.32NTDD281 pKa = 3.79DD282 pKa = 6.02SNDD285 pKa = 3.79DD286 pKa = 3.47QPSARR291 pKa = 11.84MADD294 pKa = 4.07DD295 pKa = 5.11DD296 pKa = 5.87SSDD299 pKa = 3.72NNQGSVTPTAYY310 pKa = 9.49TPATNSDD317 pKa = 3.62GGSQVTPASTPAATNRR333 pKa = 11.84VNTEE337 pKa = 3.51ALINWFYY344 pKa = 11.81NHH346 pKa = 6.4MGKK349 pKa = 8.26LTYY352 pKa = 11.2DD353 pKa = 3.12MGGSRR358 pKa = 11.84NGSDD362 pKa = 3.01GTADD366 pKa = 3.58CSGSMTEE373 pKa = 4.5ALYY376 pKa = 10.53EE377 pKa = 4.27AGASKK382 pKa = 9.89PAYY385 pKa = 10.14LYY387 pKa = 9.76STEE390 pKa = 4.31NLHH393 pKa = 7.53SYY395 pKa = 7.97LTQNGYY401 pKa = 9.86HH402 pKa = 6.43LVAVDD407 pKa = 4.31QPWDD411 pKa = 3.52AQRR414 pKa = 11.84GDD416 pKa = 3.66VVIWGQKK423 pKa = 8.47GASLGSAGHH432 pKa = 5.83AQVMVDD438 pKa = 3.34QNNAISVNYY447 pKa = 10.25AHH449 pKa = 7.62GEE451 pKa = 4.0QQGAAVSVWNYY462 pKa = 10.38ANAYY466 pKa = 10.07AYY468 pKa = 10.18NQEE471 pKa = 4.23ANGGHH476 pKa = 5.83LTYY479 pKa = 10.62YY480 pKa = 10.52VYY482 pKa = 10.63RR483 pKa = 11.84QQ484 pKa = 3.09
MM1 pKa = 7.71EE2 pKa = 4.86NQTMKK7 pKa = 10.69QLSKK11 pKa = 11.11RR12 pKa = 11.84LMVTAGASTLAATGAVLGATVVGNAQTASANDD44 pKa = 3.4VTEE47 pKa = 4.39PTSQTDD53 pKa = 4.0AQNQWKK59 pKa = 9.94ANSVDD64 pKa = 3.52HH65 pKa = 6.59VKK67 pKa = 10.36QAVKK71 pKa = 10.24KK72 pKa = 10.53QSAKK76 pKa = 10.42DD77 pKa = 3.51LKK79 pKa = 10.69DD80 pKa = 3.82YY81 pKa = 10.37QVQWGDD87 pKa = 3.52TLSAIAEE94 pKa = 4.35AFGTTADD101 pKa = 4.23AAGQQLGLSDD111 pKa = 4.5HH112 pKa = 6.6GLLVAGQTMGKK123 pKa = 8.51QEE125 pKa = 4.88EE126 pKa = 4.6IIQGLKK132 pKa = 7.65TAGYY136 pKa = 9.43LADD139 pKa = 4.18GQQTQQADD147 pKa = 3.79LGKK150 pKa = 10.54GDD152 pKa = 3.71QNLIMVQQGTPTLDD166 pKa = 3.45TNNGQQTTEE175 pKa = 3.7NTTVNNQTNSQSGSQAAYY193 pKa = 9.3EE194 pKa = 4.2QASQAAAEE202 pKa = 4.28SQAISQSNSMAAATAATNQYY222 pKa = 8.85SQSAASQANNDD233 pKa = 3.46ASQASLTADD242 pKa = 3.4SEE244 pKa = 4.96SQVTPAASDD253 pKa = 3.42ATSTSDD259 pKa = 4.21DD260 pKa = 3.67AAATTPASDD269 pKa = 3.55SSASNSDD276 pKa = 3.14SDD278 pKa = 4.32NTDD281 pKa = 3.79DD282 pKa = 6.02SNDD285 pKa = 3.79DD286 pKa = 3.47QPSARR291 pKa = 11.84MADD294 pKa = 4.07DD295 pKa = 5.11DD296 pKa = 5.87SSDD299 pKa = 3.72NNQGSVTPTAYY310 pKa = 9.49TPATNSDD317 pKa = 3.62GGSQVTPASTPAATNRR333 pKa = 11.84VNTEE337 pKa = 3.51ALINWFYY344 pKa = 11.81NHH346 pKa = 6.4MGKK349 pKa = 8.26LTYY352 pKa = 11.2DD353 pKa = 3.12MGGSRR358 pKa = 11.84NGSDD362 pKa = 3.01GTADD366 pKa = 3.58CSGSMTEE373 pKa = 4.5ALYY376 pKa = 10.53EE377 pKa = 4.27AGASKK382 pKa = 9.89PAYY385 pKa = 10.14LYY387 pKa = 9.76STEE390 pKa = 4.31NLHH393 pKa = 7.53SYY395 pKa = 7.97LTQNGYY401 pKa = 9.86HH402 pKa = 6.43LVAVDD407 pKa = 4.31QPWDD411 pKa = 3.52AQRR414 pKa = 11.84GDD416 pKa = 3.66VVIWGQKK423 pKa = 8.47GASLGSAGHH432 pKa = 5.83AQVMVDD438 pKa = 3.34QNNAISVNYY447 pKa = 10.25AHH449 pKa = 7.62GEE451 pKa = 4.0QQGAAVSVWNYY462 pKa = 10.38ANAYY466 pKa = 10.07AYY468 pKa = 10.18NQEE471 pKa = 4.23ANGGHH476 pKa = 5.83LTYY479 pKa = 10.62YY480 pKa = 10.52VYY482 pKa = 10.63RR483 pKa = 11.84QQ484 pKa = 3.09
Molecular weight: 50.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I1HNL8|A0A1I1HNL8_9LACO Helix-turn-helix domain-containing protein OS=Fructobacillus durionis OX=283737 GN=SAMN05660453_0018 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.6RR3 pKa = 11.84TFQPKK8 pKa = 8.02KK9 pKa = 7.4RR10 pKa = 11.84HH11 pKa = 5.36RR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 5.84GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MLTSNGRR28 pKa = 11.84KK29 pKa = 8.63VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.6RR3 pKa = 11.84TFQPKK8 pKa = 8.02KK9 pKa = 7.4RR10 pKa = 11.84HH11 pKa = 5.36RR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 5.84GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MLTSNGRR28 pKa = 11.84KK29 pKa = 8.63VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
394407 |
34 |
2816 |
315.5 |
34.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.274 ± 0.092 | 0.248 ± 0.013 |
6.155 ± 0.069 | 6.02 ± 0.08 |
4.264 ± 0.056 | 6.917 ± 0.063 |
1.825 ± 0.027 | 6.269 ± 0.066 |
6.726 ± 0.062 | 9.781 ± 0.09 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.634 ± 0.032 | 4.436 ± 0.06 |
3.507 ± 0.042 | 4.517 ± 0.061 |
3.856 ± 0.056 | 6.046 ± 0.076 |
5.756 ± 0.067 | 7.419 ± 0.056 |
1.048 ± 0.027 | 3.301 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
