
Dragonfly larvae associated circular virus-2
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.01
Get precalculated fractions of proteins
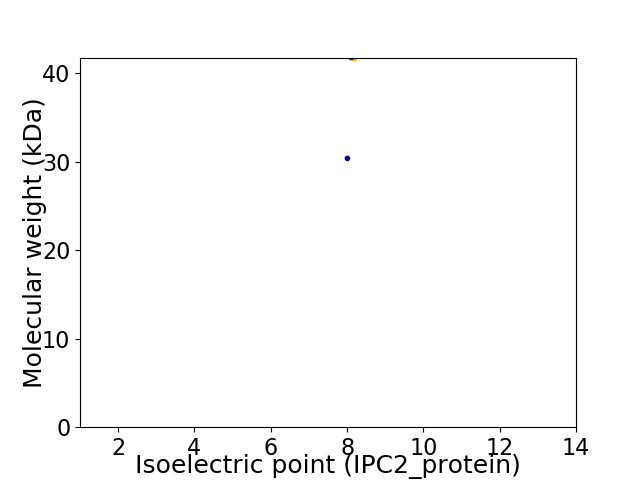
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
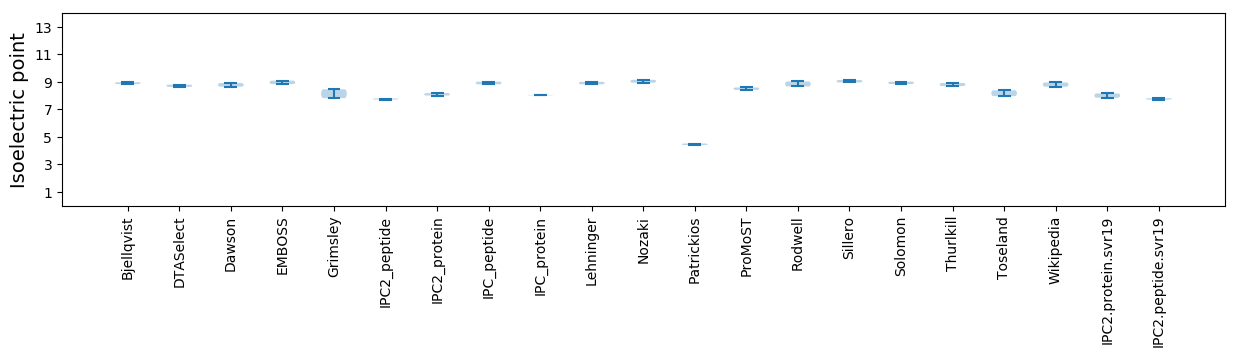
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5U7Q2|W5U7Q2_9VIRU Replication-associated protein OS=Dragonfly larvae associated circular virus-2 OX=1454023 PE=4 SV=1
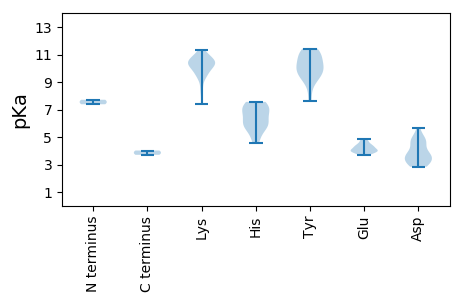
MM1 pKa = 7.71ARR3 pKa = 11.84RR4 pKa = 11.84QGIFWLLTIPRR15 pKa = 11.84HH16 pKa = 5.03EE17 pKa = 4.45FVPYY21 pKa = 10.54LPPGCVWIRR30 pKa = 11.84GQLEE34 pKa = 3.87EE35 pKa = 4.79GGEE38 pKa = 4.06DD39 pKa = 5.65GYY41 pKa = 10.82LHH43 pKa = 6.0WQVMVAFTKK52 pKa = 10.54KK53 pKa = 9.89IGLSGVKK60 pKa = 10.2VLFGPAAHH68 pKa = 7.05AEE70 pKa = 4.19LSRR73 pKa = 11.84SAAATQYY80 pKa = 9.33VWKK83 pKa = 10.38EE84 pKa = 3.9EE85 pKa = 3.94TRR87 pKa = 11.84VAGTQFNLGAKK98 pKa = 9.51PFEE101 pKa = 4.68RR102 pKa = 11.84NSKK105 pKa = 10.23RR106 pKa = 11.84DD107 pKa = 2.95WDD109 pKa = 4.66SIWLSAQEE117 pKa = 4.51GNLEE121 pKa = 4.87AIPADD126 pKa = 3.44VRR128 pKa = 11.84VVSYY132 pKa = 8.54RR133 pKa = 11.84TLRR136 pKa = 11.84AIGSDD141 pKa = 3.69YY142 pKa = 11.17AVPRR146 pKa = 11.84ALEE149 pKa = 4.0RR150 pKa = 11.84TCYY153 pKa = 10.23VFWGSTGTGKK163 pKa = 10.38SRR165 pKa = 11.84TAWEE169 pKa = 4.0QAGMDD174 pKa = 4.85AYY176 pKa = 11.26CKK178 pKa = 10.38DD179 pKa = 3.65PRR181 pKa = 11.84SKK183 pKa = 10.54FWDD186 pKa = 5.21GYY188 pKa = 9.86CSQEE192 pKa = 3.68NVVIDD197 pKa = 3.95EE198 pKa = 4.19FRR200 pKa = 11.84GGIDD204 pKa = 2.84ISHH207 pKa = 7.48ILRR210 pKa = 11.84WLDD213 pKa = 3.42RR214 pKa = 11.84YY215 pKa = 9.28PVRR218 pKa = 11.84VEE220 pKa = 3.72IKK222 pKa = 10.01GSSRR226 pKa = 11.84PLQATKK232 pKa = 10.67LWITSNLDD240 pKa = 3.26PNLWYY245 pKa = 9.87PDD247 pKa = 3.04IDD249 pKa = 4.69AEE251 pKa = 4.35TLLALRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84LTITHH264 pKa = 6.96FAA266 pKa = 4.02
MM1 pKa = 7.71ARR3 pKa = 11.84RR4 pKa = 11.84QGIFWLLTIPRR15 pKa = 11.84HH16 pKa = 5.03EE17 pKa = 4.45FVPYY21 pKa = 10.54LPPGCVWIRR30 pKa = 11.84GQLEE34 pKa = 3.87EE35 pKa = 4.79GGEE38 pKa = 4.06DD39 pKa = 5.65GYY41 pKa = 10.82LHH43 pKa = 6.0WQVMVAFTKK52 pKa = 10.54KK53 pKa = 9.89IGLSGVKK60 pKa = 10.2VLFGPAAHH68 pKa = 7.05AEE70 pKa = 4.19LSRR73 pKa = 11.84SAAATQYY80 pKa = 9.33VWKK83 pKa = 10.38EE84 pKa = 3.9EE85 pKa = 3.94TRR87 pKa = 11.84VAGTQFNLGAKK98 pKa = 9.51PFEE101 pKa = 4.68RR102 pKa = 11.84NSKK105 pKa = 10.23RR106 pKa = 11.84DD107 pKa = 2.95WDD109 pKa = 4.66SIWLSAQEE117 pKa = 4.51GNLEE121 pKa = 4.87AIPADD126 pKa = 3.44VRR128 pKa = 11.84VVSYY132 pKa = 8.54RR133 pKa = 11.84TLRR136 pKa = 11.84AIGSDD141 pKa = 3.69YY142 pKa = 11.17AVPRR146 pKa = 11.84ALEE149 pKa = 4.0RR150 pKa = 11.84TCYY153 pKa = 10.23VFWGSTGTGKK163 pKa = 10.38SRR165 pKa = 11.84TAWEE169 pKa = 4.0QAGMDD174 pKa = 4.85AYY176 pKa = 11.26CKK178 pKa = 10.38DD179 pKa = 3.65PRR181 pKa = 11.84SKK183 pKa = 10.54FWDD186 pKa = 5.21GYY188 pKa = 9.86CSQEE192 pKa = 3.68NVVIDD197 pKa = 3.95EE198 pKa = 4.19FRR200 pKa = 11.84GGIDD204 pKa = 2.84ISHH207 pKa = 7.48ILRR210 pKa = 11.84WLDD213 pKa = 3.42RR214 pKa = 11.84YY215 pKa = 9.28PVRR218 pKa = 11.84VEE220 pKa = 3.72IKK222 pKa = 10.01GSSRR226 pKa = 11.84PLQATKK232 pKa = 10.67LWITSNLDD240 pKa = 3.26PNLWYY245 pKa = 9.87PDD247 pKa = 3.04IDD249 pKa = 4.69AEE251 pKa = 4.35TLLALRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84LTITHH264 pKa = 6.96FAA266 pKa = 4.02
Molecular weight: 30.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5U7Q2|W5U7Q2_9VIRU Replication-associated protein OS=Dragonfly larvae associated circular virus-2 OX=1454023 PE=4 SV=1
MM1 pKa = 7.38TKK3 pKa = 10.47LHH5 pKa = 6.05VPYY8 pKa = 7.6MTPYY12 pKa = 10.88RR13 pKa = 11.84NGTSQSHH20 pKa = 5.7GKK22 pKa = 9.76RR23 pKa = 11.84FKK25 pKa = 11.33LEE27 pKa = 3.87VKK29 pKa = 10.45GEE31 pKa = 4.02YY32 pKa = 10.04KK33 pKa = 9.43RR34 pKa = 11.84THH36 pKa = 5.7SQTTTEE42 pKa = 3.98KK43 pKa = 10.97AEE45 pKa = 4.0EE46 pKa = 4.09RR47 pKa = 11.84MHH49 pKa = 7.55GDD51 pKa = 3.7DD52 pKa = 3.81VQSGITGTGMSFPGHH67 pKa = 5.92SRR69 pKa = 11.84HH70 pKa = 6.73KK71 pKa = 10.79DD72 pKa = 3.24VLRR75 pKa = 11.84YY76 pKa = 9.63AQVKK80 pKa = 9.74NNEE83 pKa = 4.25TYY85 pKa = 11.22GLIVDD90 pKa = 4.93CVAGSQATVTLATVGNTEE108 pKa = 3.79QCIGKK113 pKa = 9.43VNPPGPFTTSLPVWKK128 pKa = 9.82AQIDD132 pKa = 3.73AATVLNSPWTASDD145 pKa = 3.48GSNNAMQKK153 pKa = 10.28INIGFCHH160 pKa = 5.85HH161 pKa = 7.2TIDD164 pKa = 4.25FTNVTNAAAVVDD176 pKa = 4.54VYY178 pKa = 11.43VVEE181 pKa = 4.66CKK183 pKa = 10.03QDD185 pKa = 3.04IPAINAGRR193 pKa = 11.84HH194 pKa = 4.58GFLGIWAHH202 pKa = 6.86GEE204 pKa = 4.08GTEE207 pKa = 4.32SGGLASQTVTTTTATLGSSSFNVVGAKK234 pKa = 8.3PTDD237 pKa = 2.9ISLFKK242 pKa = 10.54KK243 pKa = 10.1YY244 pKa = 11.04YY245 pKa = 9.82KK246 pKa = 10.12VLKK249 pKa = 8.58AHH251 pKa = 6.52CMHH254 pKa = 7.42LGPGATEE261 pKa = 3.95VVNISSACNVDD272 pKa = 3.4VDD274 pKa = 4.03IEE276 pKa = 4.33KK277 pKa = 11.04YY278 pKa = 9.4LAKK281 pKa = 10.9NNVLVPDD288 pKa = 4.56LADD291 pKa = 2.97TAMKK295 pKa = 9.36IWAMKK300 pKa = 10.35GSFEE304 pKa = 4.38IYY306 pKa = 10.18CIARR310 pKa = 11.84GTAMLDD316 pKa = 3.54TNVLDD321 pKa = 4.8LANTTTAPVKK331 pKa = 10.33LAVVVQRR338 pKa = 11.84KK339 pKa = 7.42YY340 pKa = 10.55QFNPMKK346 pKa = 10.5YY347 pKa = 9.22KK348 pKa = 10.56SKK350 pKa = 10.58SFKK353 pKa = 10.93AVVGYY358 pKa = 10.21QKK360 pKa = 11.06LPFGVPAANLKK371 pKa = 9.24TEE373 pKa = 4.36GPTANISVAFTAAAA387 pKa = 3.71
MM1 pKa = 7.38TKK3 pKa = 10.47LHH5 pKa = 6.05VPYY8 pKa = 7.6MTPYY12 pKa = 10.88RR13 pKa = 11.84NGTSQSHH20 pKa = 5.7GKK22 pKa = 9.76RR23 pKa = 11.84FKK25 pKa = 11.33LEE27 pKa = 3.87VKK29 pKa = 10.45GEE31 pKa = 4.02YY32 pKa = 10.04KK33 pKa = 9.43RR34 pKa = 11.84THH36 pKa = 5.7SQTTTEE42 pKa = 3.98KK43 pKa = 10.97AEE45 pKa = 4.0EE46 pKa = 4.09RR47 pKa = 11.84MHH49 pKa = 7.55GDD51 pKa = 3.7DD52 pKa = 3.81VQSGITGTGMSFPGHH67 pKa = 5.92SRR69 pKa = 11.84HH70 pKa = 6.73KK71 pKa = 10.79DD72 pKa = 3.24VLRR75 pKa = 11.84YY76 pKa = 9.63AQVKK80 pKa = 9.74NNEE83 pKa = 4.25TYY85 pKa = 11.22GLIVDD90 pKa = 4.93CVAGSQATVTLATVGNTEE108 pKa = 3.79QCIGKK113 pKa = 9.43VNPPGPFTTSLPVWKK128 pKa = 9.82AQIDD132 pKa = 3.73AATVLNSPWTASDD145 pKa = 3.48GSNNAMQKK153 pKa = 10.28INIGFCHH160 pKa = 5.85HH161 pKa = 7.2TIDD164 pKa = 4.25FTNVTNAAAVVDD176 pKa = 4.54VYY178 pKa = 11.43VVEE181 pKa = 4.66CKK183 pKa = 10.03QDD185 pKa = 3.04IPAINAGRR193 pKa = 11.84HH194 pKa = 4.58GFLGIWAHH202 pKa = 6.86GEE204 pKa = 4.08GTEE207 pKa = 4.32SGGLASQTVTTTTATLGSSSFNVVGAKK234 pKa = 8.3PTDD237 pKa = 2.9ISLFKK242 pKa = 10.54KK243 pKa = 10.1YY244 pKa = 11.04YY245 pKa = 9.82KK246 pKa = 10.12VLKK249 pKa = 8.58AHH251 pKa = 6.52CMHH254 pKa = 7.42LGPGATEE261 pKa = 3.95VVNISSACNVDD272 pKa = 3.4VDD274 pKa = 4.03IEE276 pKa = 4.33KK277 pKa = 11.04YY278 pKa = 9.4LAKK281 pKa = 10.9NNVLVPDD288 pKa = 4.56LADD291 pKa = 2.97TAMKK295 pKa = 9.36IWAMKK300 pKa = 10.35GSFEE304 pKa = 4.38IYY306 pKa = 10.18CIARR310 pKa = 11.84GTAMLDD316 pKa = 3.54TNVLDD321 pKa = 4.8LANTTTAPVKK331 pKa = 10.33LAVVVQRR338 pKa = 11.84KK339 pKa = 7.42YY340 pKa = 10.55QFNPMKK346 pKa = 10.5YY347 pKa = 9.22KK348 pKa = 10.56SKK350 pKa = 10.58SFKK353 pKa = 10.93AVVGYY358 pKa = 10.21QKK360 pKa = 11.06LPFGVPAANLKK371 pKa = 9.24TEE373 pKa = 4.36GPTANISVAFTAAAA387 pKa = 3.71
Molecular weight: 41.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
653 |
266 |
387 |
326.5 |
36.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.495 ± 0.547 | 1.685 ± 0.117 |
4.594 ± 0.432 | 4.594 ± 0.917 |
3.522 ± 0.153 | 7.963 ± 0.044 |
2.603 ± 0.467 | 4.9 ± 0.476 |
6.126 ± 1.284 | 6.891 ± 1.132 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.991 ± 0.557 | 4.288 ± 1.311 |
4.594 ± 0.189 | 3.369 ± 0.009 |
4.9 ± 2.417 | 5.972 ± 0.027 |
8.27 ± 1.697 | 8.27 ± 1.212 |
2.45 ± 1.33 | 3.522 ± 0.153 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
