
Sweet potato leaf curl Spain virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus; unclassified Begomovirus
Average proteome isoelectric point is 7.47
Get precalculated fractions of proteins
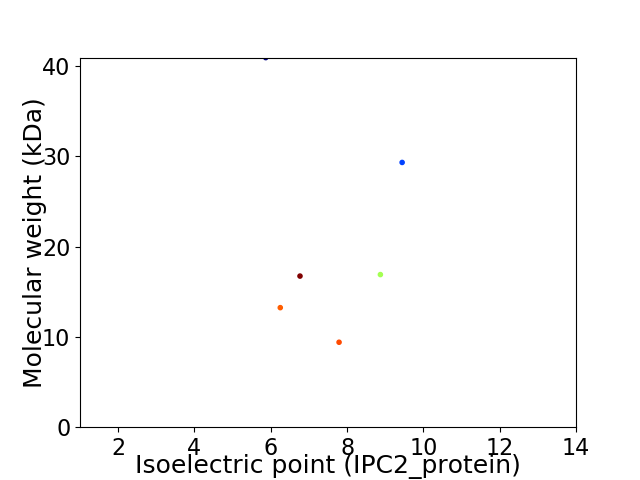
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I6LI28|I6LI28_9GEMI Replication-associated protein OS=Sweet potato leaf curl Spain virus OX=652717 GN=C1 PE=3 SV=1
MM1 pKa = 7.31PRR3 pKa = 11.84AGRR6 pKa = 11.84FNINAKK12 pKa = 10.36NYY14 pKa = 9.14FLTYY18 pKa = 7.87PQCSISKK25 pKa = 10.05EE26 pKa = 3.79EE27 pKa = 4.29ALAQILNIPTAVNKK41 pKa = 10.32KK42 pKa = 9.63FIKK45 pKa = 9.69ICRR48 pKa = 11.84EE49 pKa = 3.76LHH51 pKa = 6.97EE52 pKa = 5.88DD53 pKa = 3.95GQPHH57 pKa = 6.66LHH59 pKa = 6.38VLLQFEE65 pKa = 5.61GKK67 pKa = 9.18FQCTNQRR74 pKa = 11.84LFDD77 pKa = 4.26LVSQTRR83 pKa = 11.84SAHH86 pKa = 4.74FHH88 pKa = 6.49PNIQRR93 pKa = 11.84AKK95 pKa = 9.77SSSDD99 pKa = 2.99VKK101 pKa = 11.22SYY103 pKa = 11.11VDD105 pKa = 3.65KK106 pKa = 11.62DD107 pKa = 3.31GDD109 pKa = 3.77TLEE112 pKa = 4.1WGEE115 pKa = 3.95FQVDD119 pKa = 3.21GRR121 pKa = 11.84SARR124 pKa = 11.84GGQQTANDD132 pKa = 3.91AAAEE136 pKa = 3.92ALNAGSKK143 pKa = 10.24DD144 pKa = 3.18AALQIIRR151 pKa = 11.84EE152 pKa = 4.06KK153 pKa = 10.91LPEE156 pKa = 3.95KK157 pKa = 10.64FIFQYY162 pKa = 11.08HH163 pKa = 5.73NLVSNLDD170 pKa = 4.36RR171 pKa = 11.84IFSPPPSVYY180 pKa = 10.41SSPFSISSFNNVPDD194 pKa = 5.62IISDD198 pKa = 3.84WAAEE202 pKa = 4.12NVMDD206 pKa = 5.32AAARR210 pKa = 11.84PEE212 pKa = 4.01RR213 pKa = 11.84PISIVIEE220 pKa = 4.71GPSRR224 pKa = 11.84MGKK227 pKa = 6.66TVWARR232 pKa = 11.84SLGPHH237 pKa = 6.71NYY239 pKa = 10.18LCGHH243 pKa = 7.38LDD245 pKa = 4.18LSPKK249 pKa = 10.09VYY251 pKa = 10.85SNSAWYY257 pKa = 10.34NVIDD261 pKa = 4.95DD262 pKa = 4.8VNPQYY267 pKa = 11.15LKK269 pKa = 10.65HH270 pKa = 6.26FKK272 pKa = 10.69EE273 pKa = 4.43FMGAQKK279 pKa = 10.56DD280 pKa = 3.62WQSNCKK286 pKa = 9.38YY287 pKa = 10.21GKK289 pKa = 9.18PVQIKK294 pKa = 10.41GGIPTIFLCNPGEE307 pKa = 4.28GSSFKK312 pKa = 10.83LWLDD316 pKa = 3.35KK317 pKa = 11.08PEE319 pKa = 4.24QEE321 pKa = 5.02ALKK324 pKa = 10.68NWAVKK329 pKa = 10.42NAVFCDD335 pKa = 3.19VDD337 pKa = 4.13SPFWIQEE344 pKa = 4.27EE345 pKa = 4.8VSHH348 pKa = 6.74SGTNTRR354 pKa = 11.84GGQEE358 pKa = 3.91EE359 pKa = 4.49PEE361 pKa = 4.34EE362 pKa = 4.15NSS364 pKa = 3.18
MM1 pKa = 7.31PRR3 pKa = 11.84AGRR6 pKa = 11.84FNINAKK12 pKa = 10.36NYY14 pKa = 9.14FLTYY18 pKa = 7.87PQCSISKK25 pKa = 10.05EE26 pKa = 3.79EE27 pKa = 4.29ALAQILNIPTAVNKK41 pKa = 10.32KK42 pKa = 9.63FIKK45 pKa = 9.69ICRR48 pKa = 11.84EE49 pKa = 3.76LHH51 pKa = 6.97EE52 pKa = 5.88DD53 pKa = 3.95GQPHH57 pKa = 6.66LHH59 pKa = 6.38VLLQFEE65 pKa = 5.61GKK67 pKa = 9.18FQCTNQRR74 pKa = 11.84LFDD77 pKa = 4.26LVSQTRR83 pKa = 11.84SAHH86 pKa = 4.74FHH88 pKa = 6.49PNIQRR93 pKa = 11.84AKK95 pKa = 9.77SSSDD99 pKa = 2.99VKK101 pKa = 11.22SYY103 pKa = 11.11VDD105 pKa = 3.65KK106 pKa = 11.62DD107 pKa = 3.31GDD109 pKa = 3.77TLEE112 pKa = 4.1WGEE115 pKa = 3.95FQVDD119 pKa = 3.21GRR121 pKa = 11.84SARR124 pKa = 11.84GGQQTANDD132 pKa = 3.91AAAEE136 pKa = 3.92ALNAGSKK143 pKa = 10.24DD144 pKa = 3.18AALQIIRR151 pKa = 11.84EE152 pKa = 4.06KK153 pKa = 10.91LPEE156 pKa = 3.95KK157 pKa = 10.64FIFQYY162 pKa = 11.08HH163 pKa = 5.73NLVSNLDD170 pKa = 4.36RR171 pKa = 11.84IFSPPPSVYY180 pKa = 10.41SSPFSISSFNNVPDD194 pKa = 5.62IISDD198 pKa = 3.84WAAEE202 pKa = 4.12NVMDD206 pKa = 5.32AAARR210 pKa = 11.84PEE212 pKa = 4.01RR213 pKa = 11.84PISIVIEE220 pKa = 4.71GPSRR224 pKa = 11.84MGKK227 pKa = 6.66TVWARR232 pKa = 11.84SLGPHH237 pKa = 6.71NYY239 pKa = 10.18LCGHH243 pKa = 7.38LDD245 pKa = 4.18LSPKK249 pKa = 10.09VYY251 pKa = 10.85SNSAWYY257 pKa = 10.34NVIDD261 pKa = 4.95DD262 pKa = 4.8VNPQYY267 pKa = 11.15LKK269 pKa = 10.65HH270 pKa = 6.26FKK272 pKa = 10.69EE273 pKa = 4.43FMGAQKK279 pKa = 10.56DD280 pKa = 3.62WQSNCKK286 pKa = 9.38YY287 pKa = 10.21GKK289 pKa = 9.18PVQIKK294 pKa = 10.41GGIPTIFLCNPGEE307 pKa = 4.28GSSFKK312 pKa = 10.83LWLDD316 pKa = 3.35KK317 pKa = 11.08PEE319 pKa = 4.24QEE321 pKa = 5.02ALKK324 pKa = 10.68NWAVKK329 pKa = 10.42NAVFCDD335 pKa = 3.19VDD337 pKa = 4.13SPFWIQEE344 pKa = 4.27EE345 pKa = 4.8VSHH348 pKa = 6.74SGTNTRR354 pKa = 11.84GGQEE358 pKa = 3.91EE359 pKa = 4.49PEE361 pKa = 4.34EE362 pKa = 4.15NSS364 pKa = 3.18
Molecular weight: 40.94 kDa
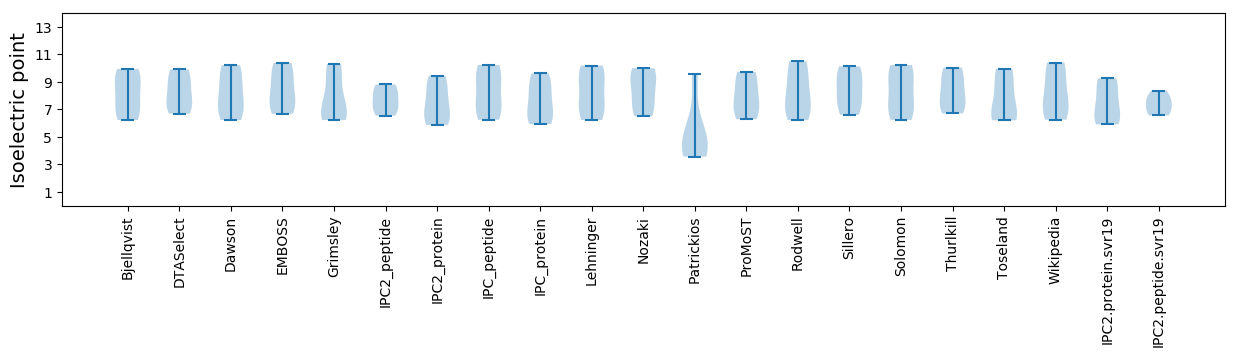
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I6LI26|I6LI26_9GEMI Replication enhancer OS=Sweet potato leaf curl Spain virus OX=652717 GN=C3 PE=3 SV=1
MM1 pKa = 7.12TGRR4 pKa = 11.84MRR6 pKa = 11.84VTRR9 pKa = 11.84RR10 pKa = 11.84FHH12 pKa = 7.1PYY14 pKa = 9.34GGTPARR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84LNFEE26 pKa = 3.95TAIVPYY32 pKa = 9.22TGNAVPIAARR42 pKa = 11.84SYY44 pKa = 10.33VPISRR49 pKa = 11.84SAQMKK54 pKa = 9.26RR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84GDD59 pKa = 4.08RR60 pKa = 11.84IPKK63 pKa = 9.96GCVGPCKK70 pKa = 10.08VQDD73 pKa = 3.86YY74 pKa = 8.72EE75 pKa = 4.55FKK77 pKa = 10.32MDD79 pKa = 3.61VPHH82 pKa = 7.03SGTFVCVSDD91 pKa = 3.96FTRR94 pKa = 11.84GTGLTHH100 pKa = 7.28RR101 pKa = 11.84LGKK104 pKa = 9.86RR105 pKa = 11.84VCIKK109 pKa = 11.05SMGIDD114 pKa = 3.27GKK116 pKa = 11.25VWMDD120 pKa = 3.8DD121 pKa = 3.32NVAKK125 pKa = 10.49GDD127 pKa = 3.52HH128 pKa = 6.23TNIITYY134 pKa = 9.02WLIRR138 pKa = 11.84DD139 pKa = 3.65RR140 pKa = 11.84RR141 pKa = 11.84PNKK144 pKa = 10.44DD145 pKa = 3.19PLTFSQAFTMYY156 pKa = 10.83DD157 pKa = 3.65NEE159 pKa = 4.15PTTAKK164 pKa = 10.17IRR166 pKa = 11.84MDD168 pKa = 3.48LRR170 pKa = 11.84DD171 pKa = 3.42RR172 pKa = 11.84MQVLKK177 pKa = 10.63KK178 pKa = 10.18FAVTVSGGPYY188 pKa = 8.51NHH190 pKa = 7.45KK191 pKa = 10.06EE192 pKa = 3.61QALVRR197 pKa = 11.84KK198 pKa = 8.56FFNSLYY204 pKa = 10.63NHH206 pKa = 5.01VTYY209 pKa = 10.84NHH211 pKa = 6.65KK212 pKa = 10.86EE213 pKa = 3.59EE214 pKa = 4.51AKK216 pKa = 10.79YY217 pKa = 10.68EE218 pKa = 4.05NHH220 pKa = 7.03LEE222 pKa = 4.07NALMLYY228 pKa = 9.88SASSHH233 pKa = 6.23ASNPVYY239 pKa = 9.46QTLRR243 pKa = 11.84CRR245 pKa = 11.84AYY247 pKa = 10.5FYY249 pKa = 10.73DD250 pKa = 3.47SHH252 pKa = 8.3KK253 pKa = 11.26NN254 pKa = 3.32
MM1 pKa = 7.12TGRR4 pKa = 11.84MRR6 pKa = 11.84VTRR9 pKa = 11.84RR10 pKa = 11.84FHH12 pKa = 7.1PYY14 pKa = 9.34GGTPARR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84LNFEE26 pKa = 3.95TAIVPYY32 pKa = 9.22TGNAVPIAARR42 pKa = 11.84SYY44 pKa = 10.33VPISRR49 pKa = 11.84SAQMKK54 pKa = 9.26RR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84GDD59 pKa = 4.08RR60 pKa = 11.84IPKK63 pKa = 9.96GCVGPCKK70 pKa = 10.08VQDD73 pKa = 3.86YY74 pKa = 8.72EE75 pKa = 4.55FKK77 pKa = 10.32MDD79 pKa = 3.61VPHH82 pKa = 7.03SGTFVCVSDD91 pKa = 3.96FTRR94 pKa = 11.84GTGLTHH100 pKa = 7.28RR101 pKa = 11.84LGKK104 pKa = 9.86RR105 pKa = 11.84VCIKK109 pKa = 11.05SMGIDD114 pKa = 3.27GKK116 pKa = 11.25VWMDD120 pKa = 3.8DD121 pKa = 3.32NVAKK125 pKa = 10.49GDD127 pKa = 3.52HH128 pKa = 6.23TNIITYY134 pKa = 9.02WLIRR138 pKa = 11.84DD139 pKa = 3.65RR140 pKa = 11.84RR141 pKa = 11.84PNKK144 pKa = 10.44DD145 pKa = 3.19PLTFSQAFTMYY156 pKa = 10.83DD157 pKa = 3.65NEE159 pKa = 4.15PTTAKK164 pKa = 10.17IRR166 pKa = 11.84MDD168 pKa = 3.48LRR170 pKa = 11.84DD171 pKa = 3.42RR172 pKa = 11.84MQVLKK177 pKa = 10.63KK178 pKa = 10.18FAVTVSGGPYY188 pKa = 8.51NHH190 pKa = 7.45KK191 pKa = 10.06EE192 pKa = 3.61QALVRR197 pKa = 11.84KK198 pKa = 8.56FFNSLYY204 pKa = 10.63NHH206 pKa = 5.01VTYY209 pKa = 10.84NHH211 pKa = 6.65KK212 pKa = 10.86EE213 pKa = 3.59EE214 pKa = 4.51AKK216 pKa = 10.79YY217 pKa = 10.68EE218 pKa = 4.05NHH220 pKa = 7.03LEE222 pKa = 4.07NALMLYY228 pKa = 9.88SASSHH233 pKa = 6.23ASNPVYY239 pKa = 9.46QTLRR243 pKa = 11.84CRR245 pKa = 11.84AYY247 pKa = 10.5FYY249 pKa = 10.73DD250 pKa = 3.47SHH252 pKa = 8.3KK253 pKa = 11.26NN254 pKa = 3.32
Molecular weight: 29.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1110 |
85 |
364 |
185.0 |
21.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.126 ± 0.513 | 2.973 ± 0.712 |
4.955 ± 0.403 | 5.405 ± 0.845 |
4.054 ± 0.548 | 5.495 ± 0.336 |
3.333 ± 0.369 | 5.586 ± 0.62 |
6.126 ± 0.344 | 7.117 ± 1.182 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.072 ± 0.574 | 5.586 ± 0.602 |
6.036 ± 0.681 | 4.414 ± 0.771 |
6.486 ± 1.176 | 8.198 ± 0.967 |
5.586 ± 1.192 | 4.865 ± 0.812 |
1.982 ± 0.319 | 3.604 ± 0.615 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
