
Aquimarina intermedia
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Aquimarina
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
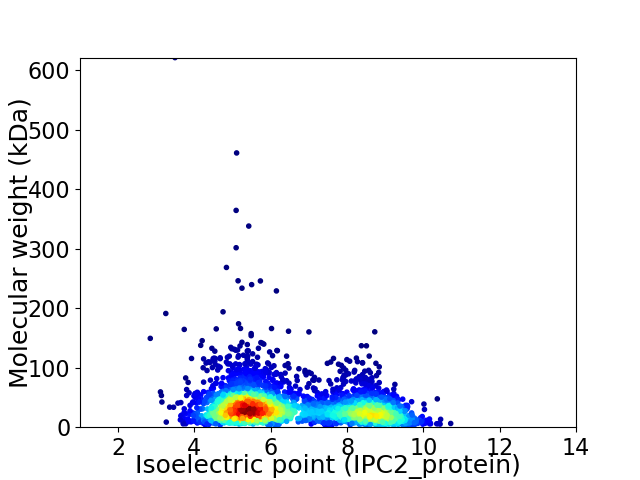
Virtual 2D-PAGE plot for 3132 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5S5C533|A0A5S5C533_9FLAO Enolase OS=Aquimarina intermedia OX=350814 GN=eno PE=3 SV=1
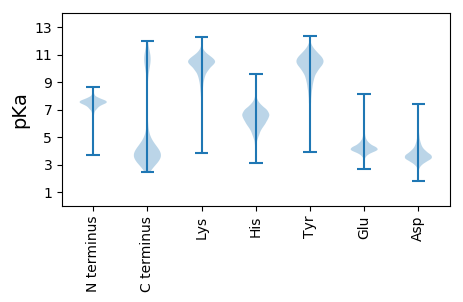
MM1 pKa = 7.66KK2 pKa = 10.25FFKK5 pKa = 9.8HH6 pKa = 5.04TFRR9 pKa = 11.84LFLLLLIIQGCNDD22 pKa = 4.51DD23 pKa = 6.06DD24 pKa = 4.97NFDD27 pKa = 3.94YY28 pKa = 11.05LNKK31 pKa = 9.84VDD33 pKa = 4.53APANIAAAIQLTQDD47 pKa = 2.93NTGLVTITPKK57 pKa = 10.94GDD59 pKa = 3.18GVVSFDD65 pKa = 4.15IYY67 pKa = 11.06FGDD70 pKa = 3.69EE71 pKa = 3.73TEE73 pKa = 4.53APVNIQLGEE82 pKa = 4.45VITHH86 pKa = 7.48AYY88 pKa = 8.6TEE90 pKa = 4.19GTYY93 pKa = 10.54DD94 pKa = 4.23LRR96 pKa = 11.84IIGIGITGLKK106 pKa = 9.02TEE108 pKa = 5.88AIQQVVISFNAPEE121 pKa = 4.03NLEE124 pKa = 4.07VVIANDD130 pKa = 3.37EE131 pKa = 4.25ATSKK135 pKa = 10.15QVNVTATADD144 pKa = 3.5YY145 pKa = 11.57AMSFDD150 pKa = 3.9VYY152 pKa = 10.69FGEE155 pKa = 4.75EE156 pKa = 4.26GNDD159 pKa = 3.42APVSATIGEE168 pKa = 4.51VASYY172 pKa = 9.08TYY174 pKa = 9.32QQPGTYY180 pKa = 8.71TIKK183 pKa = 10.69VVAKK187 pKa = 9.38GGAIATTEE195 pKa = 3.86YY196 pKa = 11.17SEE198 pKa = 4.27VFEE201 pKa = 4.34VTEE204 pKa = 3.79ILQPVASAPTPPARR218 pKa = 11.84AAQDD222 pKa = 3.43VMSVFSAAYY231 pKa = 8.14TDD233 pKa = 4.22EE234 pKa = 5.27PGTDD238 pKa = 3.88YY239 pKa = 11.16FPDD242 pKa = 3.48WGQGSQGSSWSLFDD256 pKa = 5.2LNGDD260 pKa = 4.61AMLQYY265 pKa = 10.94INLSYY270 pKa = 10.81QGIQFGAAVDD280 pKa = 3.87VSAMEE285 pKa = 4.47FFHH288 pKa = 6.43MDD290 pKa = 2.32VWTSGDD296 pKa = 3.18VTAIEE301 pKa = 4.44TSLISQTNGEE311 pKa = 4.54KK312 pKa = 10.47PFSNDD317 pKa = 2.58LVAGQWTAIDD327 pKa = 3.73IPISAFTDD335 pKa = 3.18QGLTVADD342 pKa = 3.28IHH344 pKa = 5.73QLKK347 pKa = 10.14FVGTPWAEE355 pKa = 3.58GTVFIDD361 pKa = 4.21NIYY364 pKa = 9.89FYY366 pKa = 10.35KK367 pKa = 10.91GSAQTVTAAPTPTVAAANVISMFSDD392 pKa = 3.9AYY394 pKa = 10.1TDD396 pKa = 3.51VTVDD400 pKa = 2.87TWRR403 pKa = 11.84TSWSDD408 pKa = 3.15AVLEE412 pKa = 4.28DD413 pKa = 3.51VTVEE417 pKa = 3.93GNATKK422 pKa = 10.24KK423 pKa = 10.51YY424 pKa = 9.79SALNFVGIEE433 pKa = 4.13TTSAPIDD440 pKa = 3.67ATAMTHH446 pKa = 5.52FHH448 pKa = 6.42TDD450 pKa = 2.33VWSDD454 pKa = 3.47DD455 pKa = 3.4FTEE458 pKa = 4.83FKK460 pKa = 10.53IKK462 pKa = 10.66LVDD465 pKa = 4.66FGADD469 pKa = 3.22GLFQGGDD476 pKa = 3.68DD477 pKa = 4.02VEE479 pKa = 4.73HH480 pKa = 7.47EE481 pKa = 4.28ITISNPAVGEE491 pKa = 4.21WVSLAIPLSDD501 pKa = 3.43FTGLTTRR508 pKa = 11.84ANIAQLIYY516 pKa = 10.6VGSPTGATTVYY527 pKa = 9.65IDD529 pKa = 3.64NVYY532 pKa = 10.36FHH534 pKa = 6.95NN535 pKa = 4.37
MM1 pKa = 7.66KK2 pKa = 10.25FFKK5 pKa = 9.8HH6 pKa = 5.04TFRR9 pKa = 11.84LFLLLLIIQGCNDD22 pKa = 4.51DD23 pKa = 6.06DD24 pKa = 4.97NFDD27 pKa = 3.94YY28 pKa = 11.05LNKK31 pKa = 9.84VDD33 pKa = 4.53APANIAAAIQLTQDD47 pKa = 2.93NTGLVTITPKK57 pKa = 10.94GDD59 pKa = 3.18GVVSFDD65 pKa = 4.15IYY67 pKa = 11.06FGDD70 pKa = 3.69EE71 pKa = 3.73TEE73 pKa = 4.53APVNIQLGEE82 pKa = 4.45VITHH86 pKa = 7.48AYY88 pKa = 8.6TEE90 pKa = 4.19GTYY93 pKa = 10.54DD94 pKa = 4.23LRR96 pKa = 11.84IIGIGITGLKK106 pKa = 9.02TEE108 pKa = 5.88AIQQVVISFNAPEE121 pKa = 4.03NLEE124 pKa = 4.07VVIANDD130 pKa = 3.37EE131 pKa = 4.25ATSKK135 pKa = 10.15QVNVTATADD144 pKa = 3.5YY145 pKa = 11.57AMSFDD150 pKa = 3.9VYY152 pKa = 10.69FGEE155 pKa = 4.75EE156 pKa = 4.26GNDD159 pKa = 3.42APVSATIGEE168 pKa = 4.51VASYY172 pKa = 9.08TYY174 pKa = 9.32QQPGTYY180 pKa = 8.71TIKK183 pKa = 10.69VVAKK187 pKa = 9.38GGAIATTEE195 pKa = 3.86YY196 pKa = 11.17SEE198 pKa = 4.27VFEE201 pKa = 4.34VTEE204 pKa = 3.79ILQPVASAPTPPARR218 pKa = 11.84AAQDD222 pKa = 3.43VMSVFSAAYY231 pKa = 8.14TDD233 pKa = 4.22EE234 pKa = 5.27PGTDD238 pKa = 3.88YY239 pKa = 11.16FPDD242 pKa = 3.48WGQGSQGSSWSLFDD256 pKa = 5.2LNGDD260 pKa = 4.61AMLQYY265 pKa = 10.94INLSYY270 pKa = 10.81QGIQFGAAVDD280 pKa = 3.87VSAMEE285 pKa = 4.47FFHH288 pKa = 6.43MDD290 pKa = 2.32VWTSGDD296 pKa = 3.18VTAIEE301 pKa = 4.44TSLISQTNGEE311 pKa = 4.54KK312 pKa = 10.47PFSNDD317 pKa = 2.58LVAGQWTAIDD327 pKa = 3.73IPISAFTDD335 pKa = 3.18QGLTVADD342 pKa = 3.28IHH344 pKa = 5.73QLKK347 pKa = 10.14FVGTPWAEE355 pKa = 3.58GTVFIDD361 pKa = 4.21NIYY364 pKa = 9.89FYY366 pKa = 10.35KK367 pKa = 10.91GSAQTVTAAPTPTVAAANVISMFSDD392 pKa = 3.9AYY394 pKa = 10.1TDD396 pKa = 3.51VTVDD400 pKa = 2.87TWRR403 pKa = 11.84TSWSDD408 pKa = 3.15AVLEE412 pKa = 4.28DD413 pKa = 3.51VTVEE417 pKa = 3.93GNATKK422 pKa = 10.24KK423 pKa = 10.51YY424 pKa = 9.79SALNFVGIEE433 pKa = 4.13TTSAPIDD440 pKa = 3.67ATAMTHH446 pKa = 5.52FHH448 pKa = 6.42TDD450 pKa = 2.33VWSDD454 pKa = 3.47DD455 pKa = 3.4FTEE458 pKa = 4.83FKK460 pKa = 10.53IKK462 pKa = 10.66LVDD465 pKa = 4.66FGADD469 pKa = 3.22GLFQGGDD476 pKa = 3.68DD477 pKa = 4.02VEE479 pKa = 4.73HH480 pKa = 7.47EE481 pKa = 4.28ITISNPAVGEE491 pKa = 4.21WVSLAIPLSDD501 pKa = 3.43FTGLTTRR508 pKa = 11.84ANIAQLIYY516 pKa = 10.6VGSPTGATTVYY527 pKa = 9.65IDD529 pKa = 3.64NVYY532 pKa = 10.36FHH534 pKa = 6.95NN535 pKa = 4.37
Molecular weight: 57.83 kDa
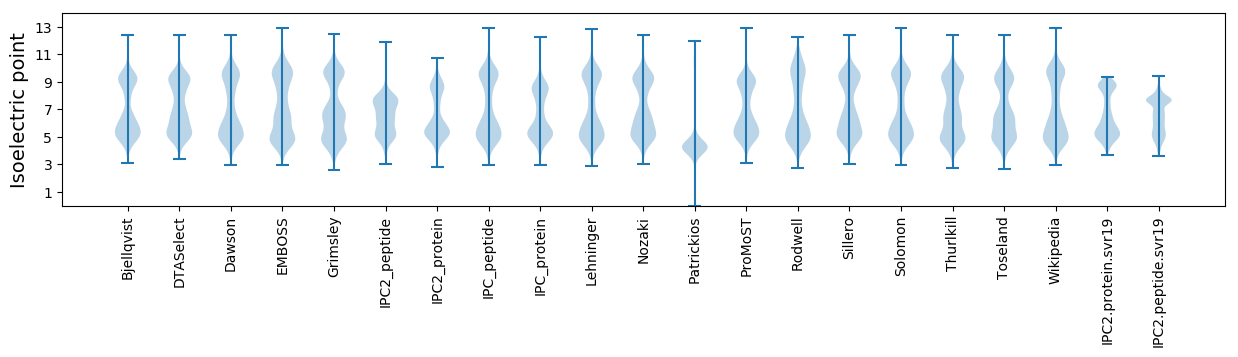
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5S5BX28|A0A5S5BX28_9FLAO Ribosomal protein S18 acetylase RimI-like enzyme OS=Aquimarina intermedia OX=350814 GN=BD809_11124 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.66LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 5.43KK51 pKa = 10.29HH52 pKa = 5.04
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.05RR21 pKa = 11.84MASVNGRR28 pKa = 11.84KK29 pKa = 9.21VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.66LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 5.43KK51 pKa = 10.29HH52 pKa = 5.04
Molecular weight: 6.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1033828 |
29 |
5960 |
330.1 |
37.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.566 ± 0.052 | 0.738 ± 0.016 |
5.686 ± 0.035 | 6.609 ± 0.048 |
5.04 ± 0.038 | 6.278 ± 0.059 |
1.863 ± 0.022 | 8.052 ± 0.045 |
7.452 ± 0.074 | 9.339 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.152 ± 0.023 | 5.686 ± 0.04 |
3.374 ± 0.03 | 3.594 ± 0.028 |
3.692 ± 0.028 | 6.481 ± 0.033 |
6.124 ± 0.069 | 6.241 ± 0.038 |
0.98 ± 0.016 | 4.053 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
