
Eggerthella lenta (strain ATCC 25559 / DSM 2243 / CCUG 17323 / JCM 9979 / KCTC 3265 / NCTC 11813 / VPI 0255 / 1899 B) (Eubacterium lentum)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Eggerthellales; Eggerthellaceae; Eggerthella; Eggerthella lenta
Average proteome isoelectric point is 5.76
Get precalculated fractions of proteins
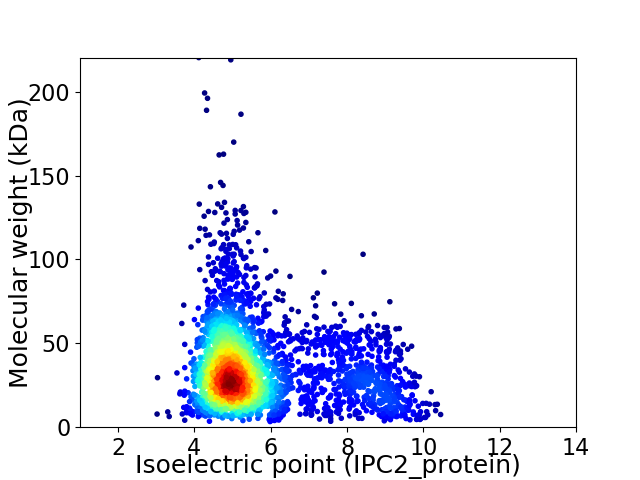
Virtual 2D-PAGE plot for 3054 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
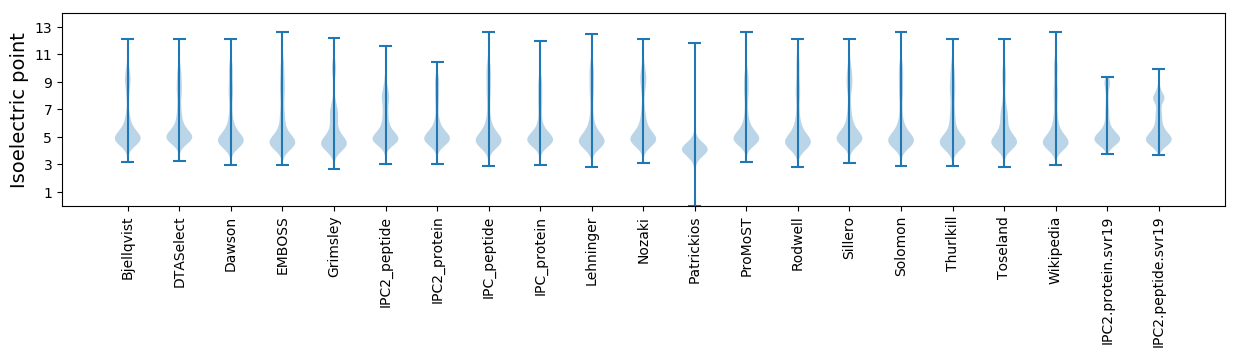
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C8WIP9|C8WIP9_EGGLE Replicative DNA helicase OS=Eggerthella lenta (strain ATCC 25559 / DSM 2243 / CCUG 17323 / JCM 9979 / KCTC 3265 / NCTC 11813 / VPI 0255 / 1899 B) OX=479437 GN=Elen_0026 PE=3 SV=1
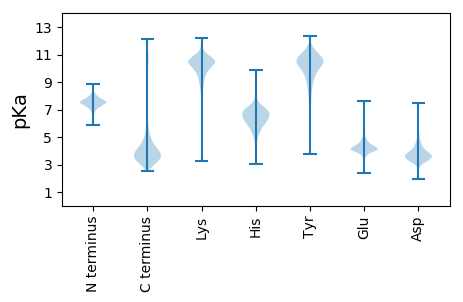
MM1 pKa = 8.05ADD3 pKa = 3.87RR4 pKa = 11.84EE5 pKa = 4.49VEE7 pKa = 4.15SSDD10 pKa = 3.72VEE12 pKa = 4.29VPEE15 pKa = 4.48IPEE18 pKa = 3.82ILEE21 pKa = 4.07KK22 pKa = 11.1VLLFSLDD29 pKa = 3.36EE30 pKa = 4.68AKK32 pKa = 10.97EE33 pKa = 3.93KK34 pKa = 8.19MTQGSDD40 pKa = 3.53VVPFTALVVKK50 pKa = 10.43EE51 pKa = 3.81NLFIEE56 pKa = 4.18NHH58 pKa = 6.28PADD61 pKa = 3.74SAEE64 pKa = 3.95EE65 pKa = 4.21CFNLARR71 pKa = 11.84HH72 pKa = 5.04TVEE75 pKa = 3.97HH76 pKa = 6.43ARR78 pKa = 11.84GAAAYY83 pKa = 8.28ALCYY87 pKa = 10.17DD88 pKa = 4.68GYY90 pKa = 10.8IEE92 pKa = 5.65IDD94 pKa = 4.06DD95 pKa = 4.26GVKK98 pKa = 10.47DD99 pKa = 3.73ALIAEE104 pKa = 4.68GGVPGEE110 pKa = 4.12DD111 pKa = 2.04TGYY114 pKa = 10.84AVSYY118 pKa = 9.52LYY120 pKa = 11.14EE121 pKa = 4.14MDD123 pKa = 3.65EE124 pKa = 4.29EE125 pKa = 5.09GNVTFEE131 pKa = 4.38EE132 pKa = 4.78EE133 pKa = 3.51PAYY136 pKa = 10.89VGEE139 pKa = 4.86APNFMIALNDD149 pKa = 3.6ADD151 pKa = 4.71SYY153 pKa = 11.43SEE155 pKa = 3.86EE156 pKa = 5.63DD157 pKa = 3.06IDD159 pKa = 4.74EE160 pKa = 4.81KK161 pKa = 11.34YY162 pKa = 11.02LEE164 pKa = 4.41EE165 pKa = 4.87DD166 pKa = 3.41AVDD169 pKa = 4.22DD170 pKa = 4.62EE171 pKa = 4.71EE172 pKa = 4.46
MM1 pKa = 8.05ADD3 pKa = 3.87RR4 pKa = 11.84EE5 pKa = 4.49VEE7 pKa = 4.15SSDD10 pKa = 3.72VEE12 pKa = 4.29VPEE15 pKa = 4.48IPEE18 pKa = 3.82ILEE21 pKa = 4.07KK22 pKa = 11.1VLLFSLDD29 pKa = 3.36EE30 pKa = 4.68AKK32 pKa = 10.97EE33 pKa = 3.93KK34 pKa = 8.19MTQGSDD40 pKa = 3.53VVPFTALVVKK50 pKa = 10.43EE51 pKa = 3.81NLFIEE56 pKa = 4.18NHH58 pKa = 6.28PADD61 pKa = 3.74SAEE64 pKa = 3.95EE65 pKa = 4.21CFNLARR71 pKa = 11.84HH72 pKa = 5.04TVEE75 pKa = 3.97HH76 pKa = 6.43ARR78 pKa = 11.84GAAAYY83 pKa = 8.28ALCYY87 pKa = 10.17DD88 pKa = 4.68GYY90 pKa = 10.8IEE92 pKa = 5.65IDD94 pKa = 4.06DD95 pKa = 4.26GVKK98 pKa = 10.47DD99 pKa = 3.73ALIAEE104 pKa = 4.68GGVPGEE110 pKa = 4.12DD111 pKa = 2.04TGYY114 pKa = 10.84AVSYY118 pKa = 9.52LYY120 pKa = 11.14EE121 pKa = 4.14MDD123 pKa = 3.65EE124 pKa = 4.29EE125 pKa = 5.09GNVTFEE131 pKa = 4.38EE132 pKa = 4.78EE133 pKa = 3.51PAYY136 pKa = 10.89VGEE139 pKa = 4.86APNFMIALNDD149 pKa = 3.6ADD151 pKa = 4.71SYY153 pKa = 11.43SEE155 pKa = 3.86EE156 pKa = 5.63DD157 pKa = 3.06IDD159 pKa = 4.74EE160 pKa = 4.81KK161 pKa = 11.34YY162 pKa = 11.02LEE164 pKa = 4.41EE165 pKa = 4.87DD166 pKa = 3.41AVDD169 pKa = 4.22DD170 pKa = 4.62EE171 pKa = 4.71EE172 pKa = 4.46
Molecular weight: 19.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C8WKC2|C8WKC2_EGGLE Transcriptional regulator AraC family OS=Eggerthella lenta (strain ATCC 25559 / DSM 2243 / CCUG 17323 / JCM 9979 / KCTC 3265 / NCTC 11813 / VPI 0255 / 1899 B) OX=479437 GN=Elen_2321 PE=4 SV=1
MM1 pKa = 7.33KK2 pKa = 10.3AVALVASEE10 pKa = 4.38GSARR14 pKa = 11.84ASEE17 pKa = 4.11ARR19 pKa = 11.84RR20 pKa = 11.84ALRR23 pKa = 11.84LDD25 pKa = 3.39PRR27 pKa = 11.84TKK29 pKa = 11.02VLMLVCANVTLLCSGFDD46 pKa = 3.2AAGFVLKK53 pKa = 10.18FLVAGIVVALLIAAGRR69 pKa = 11.84RR70 pKa = 11.84AAGIGFAAVFAAAALLEE87 pKa = 4.03QMNEE91 pKa = 3.87RR92 pKa = 11.84GLLDD96 pKa = 3.85ALGSTSAAAVTLRR109 pKa = 11.84FLSALVLQLMPGTMFAYY126 pKa = 10.68YY127 pKa = 10.6LFATTKK133 pKa = 9.9VSEE136 pKa = 4.68FVAAMEE142 pKa = 4.51RR143 pKa = 11.84VRR145 pKa = 11.84LPQRR149 pKa = 11.84VIIPFAVVFRR159 pKa = 11.84FFPTVLEE166 pKa = 4.43EE167 pKa = 3.87YY168 pKa = 10.52RR169 pKa = 11.84SIRR172 pKa = 11.84DD173 pKa = 3.36AMRR176 pKa = 11.84LRR178 pKa = 11.84GVGWRR183 pKa = 11.84SGPVALVEE191 pKa = 4.07YY192 pKa = 10.45RR193 pKa = 11.84LVPLVAGMVKK203 pKa = 10.04IGDD206 pKa = 4.04EE207 pKa = 4.07LSAASVTRR215 pKa = 11.84GLGGEE220 pKa = 4.04AVRR223 pKa = 11.84TSRR226 pKa = 11.84CRR228 pKa = 11.84IGFAAADD235 pKa = 3.52AALATVFLACAAATAAKK252 pKa = 10.66GLMGWW257 pKa = 3.62
MM1 pKa = 7.33KK2 pKa = 10.3AVALVASEE10 pKa = 4.38GSARR14 pKa = 11.84ASEE17 pKa = 4.11ARR19 pKa = 11.84RR20 pKa = 11.84ALRR23 pKa = 11.84LDD25 pKa = 3.39PRR27 pKa = 11.84TKK29 pKa = 11.02VLMLVCANVTLLCSGFDD46 pKa = 3.2AAGFVLKK53 pKa = 10.18FLVAGIVVALLIAAGRR69 pKa = 11.84RR70 pKa = 11.84AAGIGFAAVFAAAALLEE87 pKa = 4.03QMNEE91 pKa = 3.87RR92 pKa = 11.84GLLDD96 pKa = 3.85ALGSTSAAAVTLRR109 pKa = 11.84FLSALVLQLMPGTMFAYY126 pKa = 10.68YY127 pKa = 10.6LFATTKK133 pKa = 9.9VSEE136 pKa = 4.68FVAAMEE142 pKa = 4.51RR143 pKa = 11.84VRR145 pKa = 11.84LPQRR149 pKa = 11.84VIIPFAVVFRR159 pKa = 11.84FFPTVLEE166 pKa = 4.43EE167 pKa = 3.87YY168 pKa = 10.52RR169 pKa = 11.84SIRR172 pKa = 11.84DD173 pKa = 3.36AMRR176 pKa = 11.84LRR178 pKa = 11.84GVGWRR183 pKa = 11.84SGPVALVEE191 pKa = 4.07YY192 pKa = 10.45RR193 pKa = 11.84LVPLVAGMVKK203 pKa = 10.04IGDD206 pKa = 4.04EE207 pKa = 4.07LSAASVTRR215 pKa = 11.84GLGGEE220 pKa = 4.04AVRR223 pKa = 11.84TSRR226 pKa = 11.84CRR228 pKa = 11.84IGFAAADD235 pKa = 3.52AALATVFLACAAATAAKK252 pKa = 10.66GLMGWW257 pKa = 3.62
Molecular weight: 27.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1043249 |
30 |
2099 |
341.6 |
37.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.314 ± 0.06 | 1.59 ± 0.02 |
6.194 ± 0.039 | 6.506 ± 0.046 |
3.925 ± 0.029 | 8.297 ± 0.042 |
1.709 ± 0.018 | 4.747 ± 0.033 |
3.668 ± 0.034 | 9.396 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.662 ± 0.021 | 2.754 ± 0.027 |
4.369 ± 0.029 | 2.809 ± 0.024 |
6.047 ± 0.053 | 5.748 ± 0.037 |
5.08 ± 0.033 | 8.226 ± 0.042 |
1.127 ± 0.017 | 2.832 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
