
Ruminococcus sp. CAG:330
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Ruminococcus; environmental samples
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
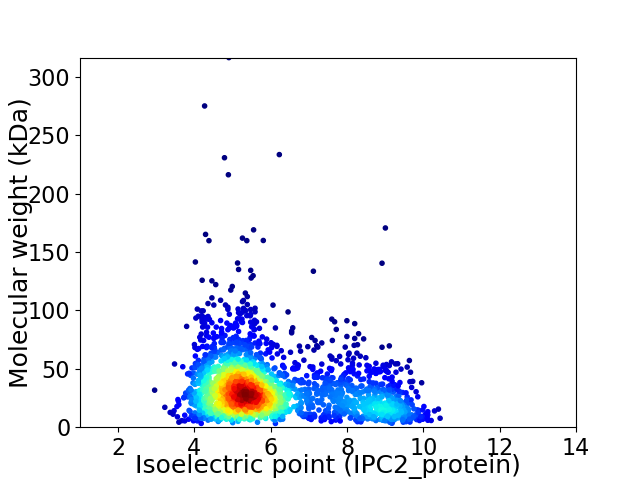
Virtual 2D-PAGE plot for 2234 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7F8N2|R7F8N2_9FIRM Uncharacterized protein OS=Ruminococcus sp. CAG:330 OX=1262954 GN=BN611_00979 PE=4 SV=1
MM1 pKa = 7.38NKK3 pKa = 9.98LKK5 pKa = 9.61KK6 pKa = 8.94TLALMAALTMTATAFVGCGDD26 pKa = 4.54KK27 pKa = 10.83EE28 pKa = 4.46DD29 pKa = 4.23SSSTADD35 pKa = 3.23SSVAEE40 pKa = 4.27SSADD44 pKa = 3.48EE45 pKa = 4.28SSADD49 pKa = 3.57AEE51 pKa = 4.37SSEE54 pKa = 4.97DD55 pKa = 3.67ADD57 pKa = 3.58SSEE60 pKa = 5.11AEE62 pKa = 4.39GGDD65 pKa = 3.47TTGAPTPDD73 pKa = 3.48GTLKK77 pKa = 11.03DD78 pKa = 4.55DD79 pKa = 5.79DD80 pKa = 4.31DD81 pKa = 4.47TLSILCWTDD90 pKa = 2.66TDD92 pKa = 4.1LKK94 pKa = 11.65AMFDD98 pKa = 3.56VTSAKK103 pKa = 10.46NPTYY107 pKa = 11.19VNVGSGGTEE116 pKa = 3.5ANEE119 pKa = 3.9QYY121 pKa = 10.82VQYY124 pKa = 9.9FSSGSDD130 pKa = 2.79VDD132 pKa = 5.81LFVCDD137 pKa = 4.45ADD139 pKa = 3.12WVMSYY144 pKa = 11.02EE145 pKa = 4.25NNDD148 pKa = 3.45EE149 pKa = 4.11YY150 pKa = 10.98TAPLSALGIDD160 pKa = 3.01EE161 pKa = 5.48SMYY164 pKa = 10.98KK165 pKa = 10.08DD166 pKa = 3.14AYY168 pKa = 10.37GYY170 pKa = 8.56TVTLGTDD177 pKa = 3.15SNGVLKK183 pKa = 10.58GASWQAAAGGYY194 pKa = 10.28AYY196 pKa = 10.52RR197 pKa = 11.84ADD199 pKa = 3.8LAEE202 pKa = 4.73EE203 pKa = 4.22YY204 pKa = 10.85LGVTSPEE211 pKa = 3.94DD212 pKa = 3.38MQAQIGDD219 pKa = 3.37WDD221 pKa = 4.47AFWKK225 pKa = 8.78TAATVYY231 pKa = 10.03EE232 pKa = 4.02KK233 pKa = 10.9SGNKK237 pKa = 8.06TAMADD242 pKa = 3.97TIAGVWRR249 pKa = 11.84AYY251 pKa = 10.45SAGNRR256 pKa = 11.84TTPWIDD262 pKa = 3.31ADD264 pKa = 5.0GKK266 pKa = 9.77FQPDD270 pKa = 3.13ATKK273 pKa = 11.09DD274 pKa = 3.87FITMAKK280 pKa = 10.04EE281 pKa = 3.99NFDD284 pKa = 3.03KK285 pKa = 11.43GYY287 pKa = 8.17ITNVKK292 pKa = 9.08QWTDD296 pKa = 2.74DD297 pKa = 3.08WKK299 pKa = 11.43LIGQSEE305 pKa = 4.42GALANATFGYY315 pKa = 9.03FFPSWSMAAGAQLQDD330 pKa = 3.63GEE332 pKa = 4.48GGEE335 pKa = 4.56GGSTYY340 pKa = 10.91GKK342 pKa = 9.98YY343 pKa = 10.53GYY345 pKa = 9.48TMGPTGWYY353 pKa = 8.51WGGSWLCVSPNCNSATAAAQFVYY376 pKa = 10.98DD377 pKa = 3.59MTINADD383 pKa = 3.4TMKK386 pKa = 10.49QYY388 pKa = 11.58ALSHH392 pKa = 6.39SDD394 pKa = 3.25FVNNKK399 pKa = 7.5TVMADD404 pKa = 3.45VVAEE408 pKa = 4.28GANKK412 pKa = 10.17NPLLKK417 pKa = 10.76DD418 pKa = 3.54GQDD421 pKa = 3.33QFATLLDD428 pKa = 3.89SADD431 pKa = 4.18NISLDD436 pKa = 3.64GVAGQNDD443 pKa = 4.16GTINDD448 pKa = 3.91AFVTAVQSYY457 pKa = 7.8CTGDD461 pKa = 3.56LDD463 pKa = 5.35SEE465 pKa = 4.54EE466 pKa = 4.79ACLDD470 pKa = 3.57NFLDD474 pKa = 4.23AVSTALPDD482 pKa = 3.7VQVDD486 pKa = 3.29
MM1 pKa = 7.38NKK3 pKa = 9.98LKK5 pKa = 9.61KK6 pKa = 8.94TLALMAALTMTATAFVGCGDD26 pKa = 4.54KK27 pKa = 10.83EE28 pKa = 4.46DD29 pKa = 4.23SSSTADD35 pKa = 3.23SSVAEE40 pKa = 4.27SSADD44 pKa = 3.48EE45 pKa = 4.28SSADD49 pKa = 3.57AEE51 pKa = 4.37SSEE54 pKa = 4.97DD55 pKa = 3.67ADD57 pKa = 3.58SSEE60 pKa = 5.11AEE62 pKa = 4.39GGDD65 pKa = 3.47TTGAPTPDD73 pKa = 3.48GTLKK77 pKa = 11.03DD78 pKa = 4.55DD79 pKa = 5.79DD80 pKa = 4.31DD81 pKa = 4.47TLSILCWTDD90 pKa = 2.66TDD92 pKa = 4.1LKK94 pKa = 11.65AMFDD98 pKa = 3.56VTSAKK103 pKa = 10.46NPTYY107 pKa = 11.19VNVGSGGTEE116 pKa = 3.5ANEE119 pKa = 3.9QYY121 pKa = 10.82VQYY124 pKa = 9.9FSSGSDD130 pKa = 2.79VDD132 pKa = 5.81LFVCDD137 pKa = 4.45ADD139 pKa = 3.12WVMSYY144 pKa = 11.02EE145 pKa = 4.25NNDD148 pKa = 3.45EE149 pKa = 4.11YY150 pKa = 10.98TAPLSALGIDD160 pKa = 3.01EE161 pKa = 5.48SMYY164 pKa = 10.98KK165 pKa = 10.08DD166 pKa = 3.14AYY168 pKa = 10.37GYY170 pKa = 8.56TVTLGTDD177 pKa = 3.15SNGVLKK183 pKa = 10.58GASWQAAAGGYY194 pKa = 10.28AYY196 pKa = 10.52RR197 pKa = 11.84ADD199 pKa = 3.8LAEE202 pKa = 4.73EE203 pKa = 4.22YY204 pKa = 10.85LGVTSPEE211 pKa = 3.94DD212 pKa = 3.38MQAQIGDD219 pKa = 3.37WDD221 pKa = 4.47AFWKK225 pKa = 8.78TAATVYY231 pKa = 10.03EE232 pKa = 4.02KK233 pKa = 10.9SGNKK237 pKa = 8.06TAMADD242 pKa = 3.97TIAGVWRR249 pKa = 11.84AYY251 pKa = 10.45SAGNRR256 pKa = 11.84TTPWIDD262 pKa = 3.31ADD264 pKa = 5.0GKK266 pKa = 9.77FQPDD270 pKa = 3.13ATKK273 pKa = 11.09DD274 pKa = 3.87FITMAKK280 pKa = 10.04EE281 pKa = 3.99NFDD284 pKa = 3.03KK285 pKa = 11.43GYY287 pKa = 8.17ITNVKK292 pKa = 9.08QWTDD296 pKa = 2.74DD297 pKa = 3.08WKK299 pKa = 11.43LIGQSEE305 pKa = 4.42GALANATFGYY315 pKa = 9.03FFPSWSMAAGAQLQDD330 pKa = 3.63GEE332 pKa = 4.48GGEE335 pKa = 4.56GGSTYY340 pKa = 10.91GKK342 pKa = 9.98YY343 pKa = 10.53GYY345 pKa = 9.48TMGPTGWYY353 pKa = 8.51WGGSWLCVSPNCNSATAAAQFVYY376 pKa = 10.98DD377 pKa = 3.59MTINADD383 pKa = 3.4TMKK386 pKa = 10.49QYY388 pKa = 11.58ALSHH392 pKa = 6.39SDD394 pKa = 3.25FVNNKK399 pKa = 7.5TVMADD404 pKa = 3.45VVAEE408 pKa = 4.28GANKK412 pKa = 10.17NPLLKK417 pKa = 10.76DD418 pKa = 3.54GQDD421 pKa = 3.33QFATLLDD428 pKa = 3.89SADD431 pKa = 4.18NISLDD436 pKa = 3.64GVAGQNDD443 pKa = 4.16GTINDD448 pKa = 3.91AFVTAVQSYY457 pKa = 7.8CTGDD461 pKa = 3.56LDD463 pKa = 5.35SEE465 pKa = 4.54EE466 pKa = 4.79ACLDD470 pKa = 3.57NFLDD474 pKa = 4.23AVSTALPDD482 pKa = 3.7VQVDD486 pKa = 3.29
Molecular weight: 51.74 kDa
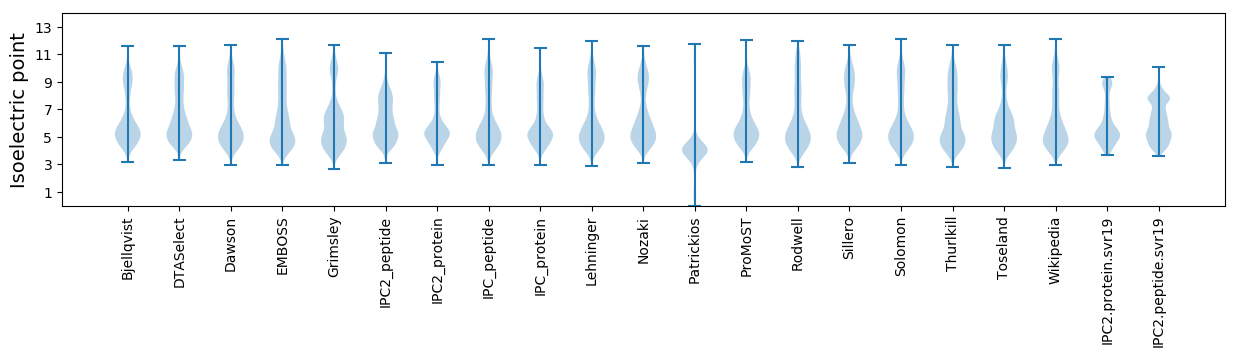
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7FBU5|R7FBU5_9FIRM Uncharacterized protein conserved in bacteria (DUF2325) OS=Ruminococcus sp. CAG:330 OX=1262954 GN=BN611_01232 PE=3 SV=1
MM1 pKa = 7.29EE2 pKa = 6.02LEE4 pKa = 4.21TFFTVPEE11 pKa = 3.97QTRR14 pKa = 11.84LLFCAVFLGIPLGLCFDD31 pKa = 4.06VLRR34 pKa = 11.84VLRR37 pKa = 11.84MLLPHH42 pKa = 6.8GKK44 pKa = 9.58LAVALEE50 pKa = 4.23DD51 pKa = 3.65TAFLLVWGGALLCFAGIFARR71 pKa = 11.84GEE73 pKa = 3.65MRR75 pKa = 11.84AYY77 pKa = 9.89YY78 pKa = 10.75ALGSTLGFLLYY89 pKa = 10.14RR90 pKa = 11.84CTIGTVTVRR99 pKa = 11.84VLHH102 pKa = 6.73RR103 pKa = 11.84IFGVSGRR110 pKa = 11.84VLRR113 pKa = 11.84WLTTPLFHH121 pKa = 6.75GVVRR125 pKa = 11.84IYY127 pKa = 11.19GVLKK131 pKa = 10.47EE132 pKa = 4.4KK133 pKa = 10.44FGHH136 pKa = 5.46FAKK139 pKa = 10.77VSGKK143 pKa = 7.59VHH145 pKa = 6.41FFWHH149 pKa = 6.52LPLIVIRR156 pKa = 11.84KK157 pKa = 8.57VLYY160 pKa = 8.42NNKK163 pKa = 9.58RR164 pKa = 11.84KK165 pKa = 7.31TVKK168 pKa = 10.28RR169 pKa = 3.71
MM1 pKa = 7.29EE2 pKa = 6.02LEE4 pKa = 4.21TFFTVPEE11 pKa = 3.97QTRR14 pKa = 11.84LLFCAVFLGIPLGLCFDD31 pKa = 4.06VLRR34 pKa = 11.84VLRR37 pKa = 11.84MLLPHH42 pKa = 6.8GKK44 pKa = 9.58LAVALEE50 pKa = 4.23DD51 pKa = 3.65TAFLLVWGGALLCFAGIFARR71 pKa = 11.84GEE73 pKa = 3.65MRR75 pKa = 11.84AYY77 pKa = 9.89YY78 pKa = 10.75ALGSTLGFLLYY89 pKa = 10.14RR90 pKa = 11.84CTIGTVTVRR99 pKa = 11.84VLHH102 pKa = 6.73RR103 pKa = 11.84IFGVSGRR110 pKa = 11.84VLRR113 pKa = 11.84WLTTPLFHH121 pKa = 6.75GVVRR125 pKa = 11.84IYY127 pKa = 11.19GVLKK131 pKa = 10.47EE132 pKa = 4.4KK133 pKa = 10.44FGHH136 pKa = 5.46FAKK139 pKa = 10.77VSGKK143 pKa = 7.59VHH145 pKa = 6.41FFWHH149 pKa = 6.52LPLIVIRR156 pKa = 11.84KK157 pKa = 8.57VLYY160 pKa = 8.42NNKK163 pKa = 9.58RR164 pKa = 11.84KK165 pKa = 7.31TVKK168 pKa = 10.28RR169 pKa = 3.71
Molecular weight: 19.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
701908 |
29 |
2801 |
314.2 |
34.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.947 ± 0.062 | 1.886 ± 0.027 |
5.644 ± 0.046 | 6.769 ± 0.053 |
3.906 ± 0.035 | 7.124 ± 0.053 |
2.013 ± 0.026 | 6.173 ± 0.05 |
5.299 ± 0.057 | 9.203 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.851 ± 0.026 | 3.737 ± 0.037 |
3.759 ± 0.033 | 4.043 ± 0.046 |
4.792 ± 0.05 | 6.037 ± 0.049 |
6.161 ± 0.086 | 6.832 ± 0.039 |
0.981 ± 0.019 | 3.839 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
