
Hydrogeniiclostidium mannosilyticum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Hydrogeniiclostidium
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
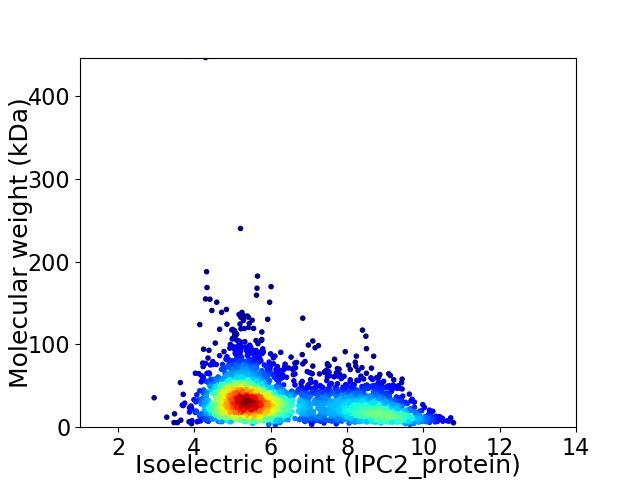
Virtual 2D-PAGE plot for 2762 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
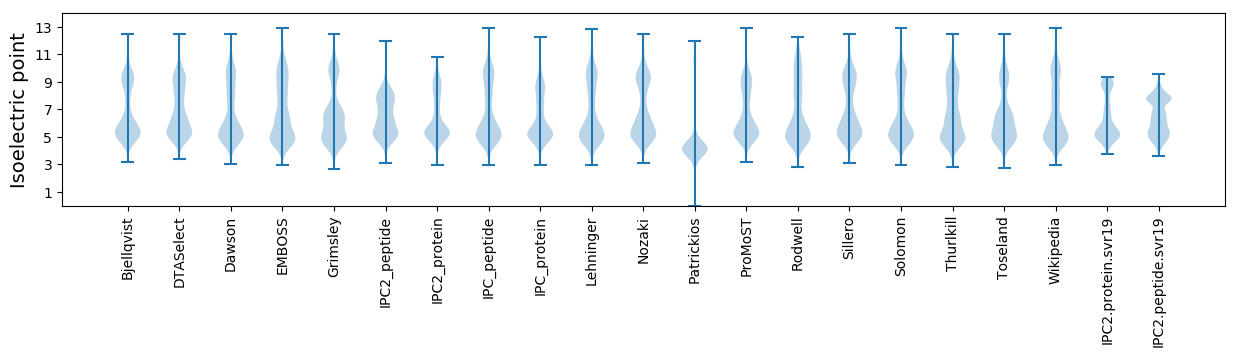
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A328UKG5|A0A328UKG5_9FIRM HTH araC/xylS-type domain-containing protein OS=Hydrogeniiclostidium mannosilyticum OX=2764322 GN=DPQ25_02840 PE=4 SV=1
MM1 pKa = 7.88FIRR4 pKa = 11.84RR5 pKa = 11.84HH6 pKa = 4.14WWAVLYY12 pKa = 8.64SVCLIAFTVYY22 pKa = 10.69LAMDD26 pKa = 3.93TFVISRR32 pKa = 11.84VYY34 pKa = 9.95TVVSEE39 pKa = 4.22TDD41 pKa = 2.81RR42 pKa = 11.84GTGSTATTDD51 pKa = 3.42TEE53 pKa = 4.07QDD55 pKa = 3.42GKK57 pKa = 11.08SDD59 pKa = 3.46SGTEE63 pKa = 4.15TVVSEE68 pKa = 4.17TSYY71 pKa = 11.49SDD73 pKa = 3.56EE74 pKa = 4.03NTQITLTEE82 pKa = 3.93YY83 pKa = 10.67RR84 pKa = 11.84EE85 pKa = 4.57CDD87 pKa = 3.07TSIYY91 pKa = 10.26VADD94 pKa = 3.7IVLSSSEE101 pKa = 3.95YY102 pKa = 10.35LQTAFAQNAYY112 pKa = 8.82GRR114 pKa = 11.84NVAEE118 pKa = 4.36KK119 pKa = 8.78TSEE122 pKa = 3.88IAEE125 pKa = 4.22SAGAILAINGDD136 pKa = 3.99YY137 pKa = 11.13YY138 pKa = 11.24GAQEE142 pKa = 4.43DD143 pKa = 5.8GYY145 pKa = 9.75VLRR148 pKa = 11.84NGVLYY153 pKa = 10.77RR154 pKa = 11.84NTAASGQEE162 pKa = 3.84DD163 pKa = 4.33LVIYY167 pKa = 10.4DD168 pKa = 5.0DD169 pKa = 4.88GSFSIINEE177 pKa = 3.97TDD179 pKa = 3.03VTAEE183 pKa = 3.91EE184 pKa = 4.28LLEE187 pKa = 6.0DD188 pKa = 4.68GAQQILSFGPALVEE202 pKa = 4.66DD203 pKa = 4.16GTVVVSEE210 pKa = 4.36DD211 pKa = 4.26DD212 pKa = 3.69EE213 pKa = 4.59VGKK216 pKa = 10.75AKK218 pKa = 10.09TSNPRR223 pKa = 11.84TAIGIIDD230 pKa = 4.14DD231 pKa = 3.71LHH233 pKa = 7.36YY234 pKa = 11.3VFVVSDD240 pKa = 3.27GRR242 pKa = 11.84TDD244 pKa = 3.29EE245 pKa = 4.52SAGLTLLQLAEE256 pKa = 4.16FMKK259 pKa = 10.75EE260 pKa = 3.91LGVTTAYY267 pKa = 10.99NLDD270 pKa = 3.92GGGSSTMYY278 pKa = 10.76FNGEE282 pKa = 4.29VINNPTTNGRR292 pKa = 11.84SIKK295 pKa = 9.64EE296 pKa = 3.84RR297 pKa = 11.84SVSDD301 pKa = 3.0IVYY304 pKa = 10.18IGYY307 pKa = 10.13
MM1 pKa = 7.88FIRR4 pKa = 11.84RR5 pKa = 11.84HH6 pKa = 4.14WWAVLYY12 pKa = 8.64SVCLIAFTVYY22 pKa = 10.69LAMDD26 pKa = 3.93TFVISRR32 pKa = 11.84VYY34 pKa = 9.95TVVSEE39 pKa = 4.22TDD41 pKa = 2.81RR42 pKa = 11.84GTGSTATTDD51 pKa = 3.42TEE53 pKa = 4.07QDD55 pKa = 3.42GKK57 pKa = 11.08SDD59 pKa = 3.46SGTEE63 pKa = 4.15TVVSEE68 pKa = 4.17TSYY71 pKa = 11.49SDD73 pKa = 3.56EE74 pKa = 4.03NTQITLTEE82 pKa = 3.93YY83 pKa = 10.67RR84 pKa = 11.84EE85 pKa = 4.57CDD87 pKa = 3.07TSIYY91 pKa = 10.26VADD94 pKa = 3.7IVLSSSEE101 pKa = 3.95YY102 pKa = 10.35LQTAFAQNAYY112 pKa = 8.82GRR114 pKa = 11.84NVAEE118 pKa = 4.36KK119 pKa = 8.78TSEE122 pKa = 3.88IAEE125 pKa = 4.22SAGAILAINGDD136 pKa = 3.99YY137 pKa = 11.13YY138 pKa = 11.24GAQEE142 pKa = 4.43DD143 pKa = 5.8GYY145 pKa = 9.75VLRR148 pKa = 11.84NGVLYY153 pKa = 10.77RR154 pKa = 11.84NTAASGQEE162 pKa = 3.84DD163 pKa = 4.33LVIYY167 pKa = 10.4DD168 pKa = 5.0DD169 pKa = 4.88GSFSIINEE177 pKa = 3.97TDD179 pKa = 3.03VTAEE183 pKa = 3.91EE184 pKa = 4.28LLEE187 pKa = 6.0DD188 pKa = 4.68GAQQILSFGPALVEE202 pKa = 4.66DD203 pKa = 4.16GTVVVSEE210 pKa = 4.36DD211 pKa = 4.26DD212 pKa = 3.69EE213 pKa = 4.59VGKK216 pKa = 10.75AKK218 pKa = 10.09TSNPRR223 pKa = 11.84TAIGIIDD230 pKa = 4.14DD231 pKa = 3.71LHH233 pKa = 7.36YY234 pKa = 11.3VFVVSDD240 pKa = 3.27GRR242 pKa = 11.84TDD244 pKa = 3.29EE245 pKa = 4.52SAGLTLLQLAEE256 pKa = 4.16FMKK259 pKa = 10.75EE260 pKa = 3.91LGVTTAYY267 pKa = 10.99NLDD270 pKa = 3.92GGGSSTMYY278 pKa = 10.76FNGEE282 pKa = 4.29VINNPTTNGRR292 pKa = 11.84SIKK295 pKa = 9.64EE296 pKa = 3.84RR297 pKa = 11.84SVSDD301 pKa = 3.0IVYY304 pKa = 10.18IGYY307 pKa = 10.13
Molecular weight: 33.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A328UEZ0|A0A328UEZ0_9FIRM Kinase to dihydroxyacetone kinase OS=Hydrogeniiclostidium mannosilyticum OX=2764322 GN=DPQ25_08355 PE=4 SV=1
MM1 pKa = 7.28GTRR4 pKa = 11.84KK5 pKa = 8.95PHH7 pKa = 6.0GLRR10 pKa = 11.84LPLCPYY16 pKa = 10.36CEE18 pKa = 3.97GRR20 pKa = 11.84FLYY23 pKa = 10.9GDD25 pKa = 3.61VRR27 pKa = 11.84RR28 pKa = 11.84HH29 pKa = 5.62IRR31 pKa = 11.84DD32 pKa = 3.31KK33 pKa = 11.35TGVCPNCGKK42 pKa = 10.44RR43 pKa = 11.84FIIRR47 pKa = 11.84AKK49 pKa = 10.51GRR51 pKa = 11.84IVLLWCIAGVLLCLLNLLFWSLEE74 pKa = 3.87QVNVLFIAGLTAVLLCCLFPLHH96 pKa = 6.56PFVVRR101 pKa = 11.84YY102 pKa = 9.53RR103 pKa = 11.84RR104 pKa = 11.84LQDD107 pKa = 3.53PLPPGRR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84GADD118 pKa = 3.01GKK120 pKa = 11.1KK121 pKa = 10.26GGKK124 pKa = 7.81TGG126 pKa = 3.31
MM1 pKa = 7.28GTRR4 pKa = 11.84KK5 pKa = 8.95PHH7 pKa = 6.0GLRR10 pKa = 11.84LPLCPYY16 pKa = 10.36CEE18 pKa = 3.97GRR20 pKa = 11.84FLYY23 pKa = 10.9GDD25 pKa = 3.61VRR27 pKa = 11.84RR28 pKa = 11.84HH29 pKa = 5.62IRR31 pKa = 11.84DD32 pKa = 3.31KK33 pKa = 11.35TGVCPNCGKK42 pKa = 10.44RR43 pKa = 11.84FIIRR47 pKa = 11.84AKK49 pKa = 10.51GRR51 pKa = 11.84IVLLWCIAGVLLCLLNLLFWSLEE74 pKa = 3.87QVNVLFIAGLTAVLLCCLFPLHH96 pKa = 6.56PFVVRR101 pKa = 11.84YY102 pKa = 9.53RR103 pKa = 11.84RR104 pKa = 11.84LQDD107 pKa = 3.53PLPPGRR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84GADD118 pKa = 3.01GKK120 pKa = 11.1KK121 pKa = 10.26GGKK124 pKa = 7.81TGG126 pKa = 3.31
Molecular weight: 14.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
860027 |
25 |
4097 |
311.4 |
34.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.281 ± 0.047 | 1.708 ± 0.018 |
5.166 ± 0.039 | 6.984 ± 0.04 |
4.144 ± 0.03 | 7.703 ± 0.037 |
1.837 ± 0.021 | 5.931 ± 0.044 |
5.297 ± 0.043 | 9.749 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.673 ± 0.022 | 3.713 ± 0.03 |
4.146 ± 0.038 | 3.664 ± 0.028 |
5.481 ± 0.049 | 5.794 ± 0.04 |
5.101 ± 0.043 | 6.806 ± 0.037 |
1.066 ± 0.02 | 3.756 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
