
Pelargonium flower break virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Alphacarmovirus
Average proteome isoelectric point is 8.7
Get precalculated fractions of proteins
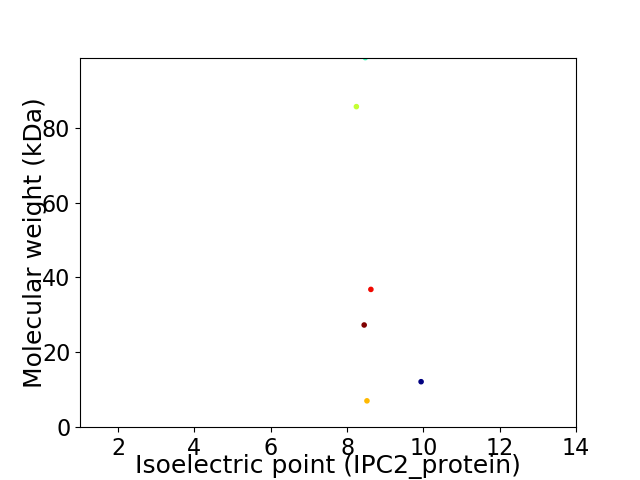
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q70WU2|Q70WU2_9TOMB RNA-directed RNA polymerase OS=Pelargonium flower break virus OX=35291 GN=p99 PE=2 SV=1
MM1 pKa = 7.41LRR3 pKa = 11.84FGSQLVVGSTLIGGLTAVGLAGLSVRR29 pKa = 11.84ATIGVVEE36 pKa = 4.53FNLRR40 pKa = 11.84CARR43 pKa = 11.84VVGDD47 pKa = 4.51IIRR50 pKa = 11.84DD51 pKa = 3.69PFNALPSAPLFPGPSVEE68 pKa = 4.28VFAEE72 pKa = 4.45EE73 pKa = 3.97VRR75 pKa = 11.84KK76 pKa = 10.25DD77 pKa = 3.67LEE79 pKa = 4.52PEE81 pKa = 4.23LEE83 pKa = 4.27AKK85 pKa = 10.32DD86 pKa = 3.55YY87 pKa = 11.48LVIHH91 pKa = 6.37EE92 pKa = 4.51EE93 pKa = 4.19KK94 pKa = 10.97EE95 pKa = 3.66MDD97 pKa = 3.55EE98 pKa = 4.17EE99 pKa = 4.74GKK101 pKa = 10.72SKK103 pKa = 10.44VVRR106 pKa = 11.84QRR108 pKa = 11.84STVNRR113 pKa = 11.84HH114 pKa = 3.8KK115 pKa = 10.65KK116 pKa = 9.55GRR118 pKa = 11.84FVHH121 pKa = 6.94RR122 pKa = 11.84LVCDD126 pKa = 4.38GKK128 pKa = 9.88NHH130 pKa = 7.3FGGTPSASRR139 pKa = 11.84ANEE142 pKa = 3.98LAVMKK147 pKa = 10.2YY148 pKa = 10.21LVGKK152 pKa = 9.96CRR154 pKa = 11.84EE155 pKa = 3.81HH156 pKa = 7.69HH157 pKa = 6.76LVVQHH162 pKa = 5.21TRR164 pKa = 11.84EE165 pKa = 4.19VCSLAMAAIFTPDD178 pKa = 2.8HH179 pKa = 6.83HH180 pKa = 6.65EE181 pKa = 3.98VNMVRR186 pKa = 11.84EE187 pKa = 4.52MNSHH191 pKa = 6.13AAYY194 pKa = 9.82RR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84VALADD202 pKa = 3.49ASRR205 pKa = 11.84VHH207 pKa = 6.22SWQQEE212 pKa = 4.15LLRR215 pKa = 11.84SPLSWNAWGRR225 pKa = 11.84AWWIANGLPDD235 pKa = 4.01RR236 pKa = 11.84EE237 pKa = 4.16PVRR240 pKa = 11.84FTKK243 pKa = 9.67XGGLFCLEE251 pKa = 4.08GVEE254 pKa = 4.24TGVRR258 pKa = 11.84RR259 pKa = 11.84GTHH262 pKa = 4.61PQMIEE267 pKa = 3.88VEE269 pKa = 4.5KK270 pKa = 11.05NSPPKK275 pKa = 10.15LRR277 pKa = 11.84KK278 pKa = 9.63LFAQRR283 pKa = 11.84LVSSGVEE290 pKa = 3.86YY291 pKa = 10.27RR292 pKa = 11.84VHH294 pKa = 5.83NHH296 pKa = 5.19SFQNLRR302 pKa = 11.84RR303 pKa = 11.84GLLEE307 pKa = 3.48RR308 pKa = 11.84VFYY311 pKa = 11.26VEE313 pKa = 4.31EE314 pKa = 4.05NKK316 pKa = 10.19QLVSCPCPEE325 pKa = 3.83KK326 pKa = 10.95GAFKK330 pKa = 10.8EE331 pKa = 3.65MGYY334 pKa = 10.13LRR336 pKa = 11.84KK337 pKa = 9.76QFVRR341 pKa = 11.84LTPKK345 pKa = 9.93HH346 pKa = 5.79ARR348 pKa = 11.84ISAKK352 pKa = 10.4EE353 pKa = 3.79FVDD356 pKa = 4.8CYY358 pKa = 10.44HH359 pKa = 6.28GRR361 pKa = 11.84KK362 pKa = 9.32KK363 pKa = 10.55SVYY366 pKa = 8.73EE367 pKa = 3.98FAAMTLSEE375 pKa = 4.62RR376 pKa = 11.84PLDD379 pKa = 4.22RR380 pKa = 11.84KK381 pKa = 10.27DD382 pKa = 4.33ASLKK386 pKa = 9.87TFIKK390 pKa = 10.68AEE392 pKa = 4.3KK393 pKa = 9.57FCKK396 pKa = 9.97ADD398 pKa = 3.44PAPRR402 pKa = 11.84VIQPRR407 pKa = 11.84SPRR410 pKa = 11.84YY411 pKa = 8.45NVEE414 pKa = 3.57LGRR417 pKa = 11.84YY418 pKa = 7.81LKK420 pKa = 10.64KK421 pKa = 10.66FEE423 pKa = 3.92HH424 pKa = 5.6TAYY427 pKa = 8.71RR428 pKa = 11.84TLDD431 pKa = 4.02KK432 pKa = 10.64IWGGKK437 pKa = 6.93TVMKK441 pKa = 10.09GYY443 pKa = 10.55SVDD446 pKa = 4.05EE447 pKa = 4.31IGKK450 pKa = 9.2HH451 pKa = 6.05ISEE454 pKa = 4.65AWDD457 pKa = 3.6SFHH460 pKa = 8.03SPVAVGFDD468 pKa = 3.23MSRR471 pKa = 11.84FDD473 pKa = 3.32QHH475 pKa = 8.03VSVPALQFEE484 pKa = 4.69HH485 pKa = 6.59SCYY488 pKa = 10.09LACFPGDD495 pKa = 3.29QHH497 pKa = 7.83LAEE500 pKa = 4.54LLSWQLKK507 pKa = 8.66NSGVGFASNGMVRR520 pKa = 11.84YY521 pKa = 9.88KK522 pKa = 10.74KK523 pKa = 10.34DD524 pKa = 3.21GCRR527 pKa = 11.84MSGDD531 pKa = 3.48MNTALGNCLLACLITRR547 pKa = 11.84HH548 pKa = 5.61LMKK551 pKa = 10.53DD552 pKa = 3.19INCRR556 pKa = 11.84LVNNGDD562 pKa = 3.73DD563 pKa = 4.12CVLFLDD569 pKa = 5.07KK570 pKa = 10.8KK571 pKa = 10.42DD572 pKa = 3.8LPFVVSNLTTGWRR585 pKa = 11.84RR586 pKa = 11.84FGFKK590 pKa = 10.26CIAEE594 pKa = 4.21EE595 pKa = 3.62PVYY598 pKa = 10.29EE599 pKa = 4.12LEE601 pKa = 4.55HH602 pKa = 7.98VRR604 pKa = 11.84FCQMAPVYY612 pKa = 10.61DD613 pKa = 4.21GSQYY617 pKa = 11.6VMLRR621 pKa = 11.84DD622 pKa = 3.9PFVSMSKK629 pKa = 10.75DD630 pKa = 3.59SFSLTHH636 pKa = 6.69WNTTKK641 pKa = 10.61NAKK644 pKa = 8.59QWMKK648 pKa = 10.86SVGICGEE655 pKa = 4.5RR656 pKa = 11.84ITGGLPVVQEE666 pKa = 4.47YY667 pKa = 9.27YY668 pKa = 10.56RR669 pKa = 11.84KK670 pKa = 8.18YY671 pKa = 10.51QEE673 pKa = 3.6IAGDD677 pKa = 3.78VKK679 pKa = 10.63FSHH682 pKa = 6.43SLEE685 pKa = 4.05ITSSGTYY692 pKa = 10.13RR693 pKa = 11.84LAQNSSRR700 pKa = 11.84AYY702 pKa = 9.95GSVSEE707 pKa = 4.18MARR710 pKa = 11.84FSFFLAFGVIPDD722 pKa = 3.77AQIALEE728 pKa = 3.9KK729 pKa = 10.0HH730 pKa = 5.63IRR732 pKa = 11.84DD733 pKa = 3.58MPVAAEE739 pKa = 4.26FGPDD743 pKa = 3.14EE744 pKa = 4.53SLADD748 pKa = 3.84HH749 pKa = 6.66TIEE752 pKa = 4.11WLLKK756 pKa = 10.01
MM1 pKa = 7.41LRR3 pKa = 11.84FGSQLVVGSTLIGGLTAVGLAGLSVRR29 pKa = 11.84ATIGVVEE36 pKa = 4.53FNLRR40 pKa = 11.84CARR43 pKa = 11.84VVGDD47 pKa = 4.51IIRR50 pKa = 11.84DD51 pKa = 3.69PFNALPSAPLFPGPSVEE68 pKa = 4.28VFAEE72 pKa = 4.45EE73 pKa = 3.97VRR75 pKa = 11.84KK76 pKa = 10.25DD77 pKa = 3.67LEE79 pKa = 4.52PEE81 pKa = 4.23LEE83 pKa = 4.27AKK85 pKa = 10.32DD86 pKa = 3.55YY87 pKa = 11.48LVIHH91 pKa = 6.37EE92 pKa = 4.51EE93 pKa = 4.19KK94 pKa = 10.97EE95 pKa = 3.66MDD97 pKa = 3.55EE98 pKa = 4.17EE99 pKa = 4.74GKK101 pKa = 10.72SKK103 pKa = 10.44VVRR106 pKa = 11.84QRR108 pKa = 11.84STVNRR113 pKa = 11.84HH114 pKa = 3.8KK115 pKa = 10.65KK116 pKa = 9.55GRR118 pKa = 11.84FVHH121 pKa = 6.94RR122 pKa = 11.84LVCDD126 pKa = 4.38GKK128 pKa = 9.88NHH130 pKa = 7.3FGGTPSASRR139 pKa = 11.84ANEE142 pKa = 3.98LAVMKK147 pKa = 10.2YY148 pKa = 10.21LVGKK152 pKa = 9.96CRR154 pKa = 11.84EE155 pKa = 3.81HH156 pKa = 7.69HH157 pKa = 6.76LVVQHH162 pKa = 5.21TRR164 pKa = 11.84EE165 pKa = 4.19VCSLAMAAIFTPDD178 pKa = 2.8HH179 pKa = 6.83HH180 pKa = 6.65EE181 pKa = 3.98VNMVRR186 pKa = 11.84EE187 pKa = 4.52MNSHH191 pKa = 6.13AAYY194 pKa = 9.82RR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84VALADD202 pKa = 3.49ASRR205 pKa = 11.84VHH207 pKa = 6.22SWQQEE212 pKa = 4.15LLRR215 pKa = 11.84SPLSWNAWGRR225 pKa = 11.84AWWIANGLPDD235 pKa = 4.01RR236 pKa = 11.84EE237 pKa = 4.16PVRR240 pKa = 11.84FTKK243 pKa = 9.67XGGLFCLEE251 pKa = 4.08GVEE254 pKa = 4.24TGVRR258 pKa = 11.84RR259 pKa = 11.84GTHH262 pKa = 4.61PQMIEE267 pKa = 3.88VEE269 pKa = 4.5KK270 pKa = 11.05NSPPKK275 pKa = 10.15LRR277 pKa = 11.84KK278 pKa = 9.63LFAQRR283 pKa = 11.84LVSSGVEE290 pKa = 3.86YY291 pKa = 10.27RR292 pKa = 11.84VHH294 pKa = 5.83NHH296 pKa = 5.19SFQNLRR302 pKa = 11.84RR303 pKa = 11.84GLLEE307 pKa = 3.48RR308 pKa = 11.84VFYY311 pKa = 11.26VEE313 pKa = 4.31EE314 pKa = 4.05NKK316 pKa = 10.19QLVSCPCPEE325 pKa = 3.83KK326 pKa = 10.95GAFKK330 pKa = 10.8EE331 pKa = 3.65MGYY334 pKa = 10.13LRR336 pKa = 11.84KK337 pKa = 9.76QFVRR341 pKa = 11.84LTPKK345 pKa = 9.93HH346 pKa = 5.79ARR348 pKa = 11.84ISAKK352 pKa = 10.4EE353 pKa = 3.79FVDD356 pKa = 4.8CYY358 pKa = 10.44HH359 pKa = 6.28GRR361 pKa = 11.84KK362 pKa = 9.32KK363 pKa = 10.55SVYY366 pKa = 8.73EE367 pKa = 3.98FAAMTLSEE375 pKa = 4.62RR376 pKa = 11.84PLDD379 pKa = 4.22RR380 pKa = 11.84KK381 pKa = 10.27DD382 pKa = 4.33ASLKK386 pKa = 9.87TFIKK390 pKa = 10.68AEE392 pKa = 4.3KK393 pKa = 9.57FCKK396 pKa = 9.97ADD398 pKa = 3.44PAPRR402 pKa = 11.84VIQPRR407 pKa = 11.84SPRR410 pKa = 11.84YY411 pKa = 8.45NVEE414 pKa = 3.57LGRR417 pKa = 11.84YY418 pKa = 7.81LKK420 pKa = 10.64KK421 pKa = 10.66FEE423 pKa = 3.92HH424 pKa = 5.6TAYY427 pKa = 8.71RR428 pKa = 11.84TLDD431 pKa = 4.02KK432 pKa = 10.64IWGGKK437 pKa = 6.93TVMKK441 pKa = 10.09GYY443 pKa = 10.55SVDD446 pKa = 4.05EE447 pKa = 4.31IGKK450 pKa = 9.2HH451 pKa = 6.05ISEE454 pKa = 4.65AWDD457 pKa = 3.6SFHH460 pKa = 8.03SPVAVGFDD468 pKa = 3.23MSRR471 pKa = 11.84FDD473 pKa = 3.32QHH475 pKa = 8.03VSVPALQFEE484 pKa = 4.69HH485 pKa = 6.59SCYY488 pKa = 10.09LACFPGDD495 pKa = 3.29QHH497 pKa = 7.83LAEE500 pKa = 4.54LLSWQLKK507 pKa = 8.66NSGVGFASNGMVRR520 pKa = 11.84YY521 pKa = 9.88KK522 pKa = 10.74KK523 pKa = 10.34DD524 pKa = 3.21GCRR527 pKa = 11.84MSGDD531 pKa = 3.48MNTALGNCLLACLITRR547 pKa = 11.84HH548 pKa = 5.61LMKK551 pKa = 10.53DD552 pKa = 3.19INCRR556 pKa = 11.84LVNNGDD562 pKa = 3.73DD563 pKa = 4.12CVLFLDD569 pKa = 5.07KK570 pKa = 10.8KK571 pKa = 10.42DD572 pKa = 3.8LPFVVSNLTTGWRR585 pKa = 11.84RR586 pKa = 11.84FGFKK590 pKa = 10.26CIAEE594 pKa = 4.21EE595 pKa = 3.62PVYY598 pKa = 10.29EE599 pKa = 4.12LEE601 pKa = 4.55HH602 pKa = 7.98VRR604 pKa = 11.84FCQMAPVYY612 pKa = 10.61DD613 pKa = 4.21GSQYY617 pKa = 11.6VMLRR621 pKa = 11.84DD622 pKa = 3.9PFVSMSKK629 pKa = 10.75DD630 pKa = 3.59SFSLTHH636 pKa = 6.69WNTTKK641 pKa = 10.61NAKK644 pKa = 8.59QWMKK648 pKa = 10.86SVGICGEE655 pKa = 4.5RR656 pKa = 11.84ITGGLPVVQEE666 pKa = 4.47YY667 pKa = 9.27YY668 pKa = 10.56RR669 pKa = 11.84KK670 pKa = 8.18YY671 pKa = 10.51QEE673 pKa = 3.6IAGDD677 pKa = 3.78VKK679 pKa = 10.63FSHH682 pKa = 6.43SLEE685 pKa = 4.05ITSSGTYY692 pKa = 10.13RR693 pKa = 11.84LAQNSSRR700 pKa = 11.84AYY702 pKa = 9.95GSVSEE707 pKa = 4.18MARR710 pKa = 11.84FSFFLAFGVIPDD722 pKa = 3.77AQIALEE728 pKa = 3.9KK729 pKa = 10.0HH730 pKa = 5.63IRR732 pKa = 11.84DD733 pKa = 3.58MPVAAEE739 pKa = 4.26FGPDD743 pKa = 3.14EE744 pKa = 4.53SLADD748 pKa = 3.84HH749 pKa = 6.66TIEE752 pKa = 4.11WLLKK756 pKa = 10.01
Molecular weight: 85.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q70WT9|Q70WT9_9TOMB Putative movement protein OS=Pelargonium flower break virus OX=35291 GN=p7 PE=2 SV=1
MM1 pKa = 8.02RR2 pKa = 11.84LTCQWEE8 pKa = 4.2DD9 pKa = 2.98VGTGVNRR16 pKa = 11.84RR17 pKa = 11.84VRR19 pKa = 11.84YY20 pKa = 8.47PSQRR24 pKa = 11.84TLSPNGHH31 pKa = 6.82LMVVMGVLGLLWLRR45 pKa = 11.84PFRR48 pKa = 11.84SIYY51 pKa = 10.21TSTFSMPPLINLQHH65 pKa = 6.61ILNLTLLSLILSSFILAEE83 pKa = 3.94RR84 pKa = 11.84VTHH87 pKa = 5.79NHH89 pKa = 5.77YY90 pKa = 11.52SNDD93 pKa = 3.43NSKK96 pKa = 10.67AQYY99 pKa = 10.29IRR101 pKa = 11.84ISTGQQ106 pKa = 2.97
MM1 pKa = 8.02RR2 pKa = 11.84LTCQWEE8 pKa = 4.2DD9 pKa = 2.98VGTGVNRR16 pKa = 11.84RR17 pKa = 11.84VRR19 pKa = 11.84YY20 pKa = 8.47PSQRR24 pKa = 11.84TLSPNGHH31 pKa = 6.82LMVVMGVLGLLWLRR45 pKa = 11.84PFRR48 pKa = 11.84SIYY51 pKa = 10.21TSTFSMPPLINLQHH65 pKa = 6.61ILNLTLLSLILSSFILAEE83 pKa = 3.94RR84 pKa = 11.84VTHH87 pKa = 5.79NHH89 pKa = 5.77YY90 pKa = 11.52SNDD93 pKa = 3.43NSKK96 pKa = 10.67AQYY99 pKa = 10.29IRR101 pKa = 11.84ISTGQQ106 pKa = 2.97
Molecular weight: 12.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2389 |
68 |
871 |
398.2 |
44.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.074 ± 0.43 | 1.967 ± 0.324 |
4.311 ± 0.231 | 5.735 ± 0.842 |
4.437 ± 0.45 | 6.949 ± 0.316 |
3.265 ± 0.526 | 3.809 ± 0.263 |
5.483 ± 0.481 | 8.707 ± 0.687 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.553 ± 0.266 | 3.809 ± 0.266 |
4.479 ± 0.195 | 2.93 ± 0.158 |
7.283 ± 0.545 | 8.204 ± 0.996 |
4.981 ± 1.271 | 9.376 ± 0.649 |
1.716 ± 0.058 | 2.805 ± 0.262 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
