
Thiorhodococcus minor
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Chromatiaceae; Thiorhodococcus
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
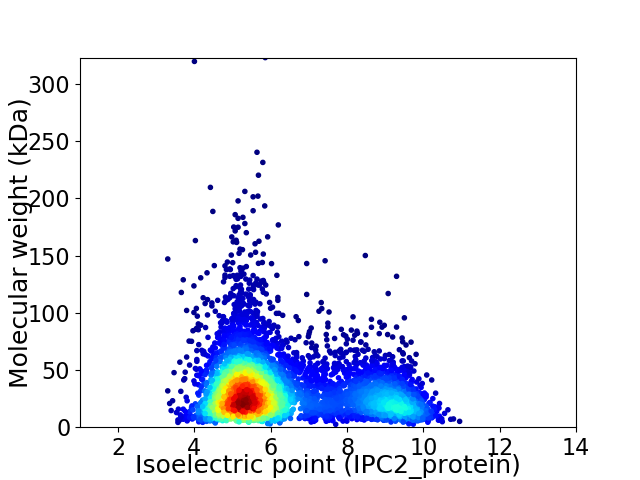
Virtual 2D-PAGE plot for 5194 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M0JXT8|A0A6M0JXT8_9GAMM Light-harvesting protein OS=Thiorhodococcus minor OX=57489 GN=G3446_06015 PE=4 SV=1
MM1 pKa = 7.75DD2 pKa = 5.01RR3 pKa = 11.84SRR5 pKa = 11.84FLSRR9 pKa = 11.84VGGGCLLAIGLAGAATPATAEE30 pKa = 3.83ISFSNVTGSSGITDD44 pKa = 3.38TGEE47 pKa = 4.03TFSTGWGDD55 pKa = 3.41FAGTGFPGIWLNKK68 pKa = 8.32HH69 pKa = 5.54QYY71 pKa = 7.86TPTRR75 pKa = 11.84IFYY78 pKa = 9.68NLSGSGFTEE87 pKa = 4.17VADD90 pKa = 3.92DD91 pKa = 3.67VLKK94 pKa = 11.23NEE96 pKa = 4.64EE97 pKa = 4.75GVPSPQTDD105 pKa = 3.86FNIPRR110 pKa = 11.84AEE112 pKa = 4.08TGPGEE117 pKa = 4.2FSYY120 pKa = 10.8FADD123 pKa = 3.37THH125 pKa = 6.05GQTSADD131 pKa = 3.45WDD133 pKa = 3.72NDD135 pKa = 2.85GDD137 pKa = 4.02QDD139 pKa = 4.02VLEE142 pKa = 4.38VTGAGYY148 pKa = 10.16IFPLWEE154 pKa = 4.05NDD156 pKa = 3.08GLGNFEE162 pKa = 4.09YY163 pKa = 10.61RR164 pKa = 11.84GDD166 pKa = 4.03EE167 pKa = 4.22LGFAYY172 pKa = 9.89PLTWEE177 pKa = 4.0RR178 pKa = 11.84RR179 pKa = 11.84EE180 pKa = 3.87RR181 pKa = 11.84PPIGGRR187 pKa = 11.84DD188 pKa = 3.54SLFFDD193 pKa = 4.16YY194 pKa = 11.05DD195 pKa = 3.55RR196 pKa = 11.84DD197 pKa = 3.56GWLDD201 pKa = 3.44AMVMARR207 pKa = 11.84EE208 pKa = 4.2NYY210 pKa = 8.86SAYY213 pKa = 8.47RR214 pKa = 11.84TPTAIFRR221 pKa = 11.84QEE223 pKa = 4.09RR224 pKa = 11.84IGGEE228 pKa = 4.1SFFTLDD234 pKa = 3.13EE235 pKa = 4.45STGVVPDD242 pKa = 4.12EE243 pKa = 4.46GTGPFPGTACEE254 pKa = 4.19FGAMAEE260 pKa = 4.06LSGDD264 pKa = 3.59SVLDD268 pKa = 4.17VICSDD273 pKa = 3.91SQISAIWDD281 pKa = 3.43VSQLPFADD289 pKa = 3.75LRR291 pKa = 11.84PVVGDD296 pKa = 3.92EE297 pKa = 3.72IFNRR301 pKa = 11.84YY302 pKa = 8.14PADD305 pKa = 3.53LAIGDD310 pKa = 4.31FNNDD314 pKa = 2.62LRR316 pKa = 11.84TDD318 pKa = 3.28IMAPTFAGADD328 pKa = 3.5YY329 pKa = 10.74VRR331 pKa = 11.84NLVGVVDD338 pKa = 4.62GQTLHH343 pKa = 6.07GWFNTSVPSNSGFNFSATGNVTFEE367 pKa = 4.18FDD369 pKa = 2.85WWTEE373 pKa = 3.84TPNIFIGADD382 pKa = 3.31GSNPDD387 pKa = 4.16LATNLPTMPFQEE399 pKa = 5.11LGHH402 pKa = 6.88GDD404 pKa = 3.78PNHH407 pKa = 5.9VVVTLSPTEE416 pKa = 4.08AQGLAPIFVPEE427 pKa = 5.18DD428 pKa = 3.46NDD430 pKa = 3.65GDD432 pKa = 4.83PITGVFIGYY441 pKa = 9.83VDD443 pKa = 3.81GEE445 pKa = 4.1WRR447 pKa = 11.84VRR449 pKa = 11.84FSGIEE454 pKa = 3.56RR455 pKa = 11.84WNVGMVITAQSGISGLTSVGGTKK478 pKa = 9.57IGQDD482 pKa = 3.07TGGNALLFLQSEE494 pKa = 4.32DD495 pKa = 3.76HH496 pKa = 6.62EE497 pKa = 4.96LVFSGGNVQAVNRR510 pKa = 11.84SCTAAAAADD519 pKa = 4.06FDD521 pKa = 4.55NDD523 pKa = 2.84MDD525 pKa = 5.34LDD527 pKa = 4.03VFFACSGQLSNLPDD541 pKa = 3.09IVYY544 pKa = 9.9EE545 pKa = 4.07NQGNGTFVALPAAGGAEE562 pKa = 3.96GQMPLGRR569 pKa = 11.84ADD571 pKa = 3.48SVMVADD577 pKa = 4.02YY578 pKa = 11.55DD579 pKa = 3.9EE580 pKa = 4.71DD581 pKa = 4.0CRR583 pKa = 11.84MDD585 pKa = 3.61VYY587 pKa = 10.09VTNGHH592 pKa = 6.1HH593 pKa = 6.29FRR595 pKa = 11.84PSSYY599 pKa = 11.24AGIHH603 pKa = 4.18QLFRR607 pKa = 11.84NTTSDD612 pKa = 3.58GNHH615 pKa = 7.35CIQIDD620 pKa = 4.01LEE622 pKa = 4.64GTVSNRR628 pKa = 11.84DD629 pKa = 3.31GIGAIVYY636 pKa = 7.54ATTPDD641 pKa = 3.19GTVQKK646 pKa = 10.36RR647 pKa = 11.84DD648 pKa = 3.55QNGGKK653 pKa = 9.2HH654 pKa = 6.14KK655 pKa = 10.44KK656 pKa = 8.6AQDD659 pKa = 3.54YY660 pKa = 10.16QRR662 pKa = 11.84VHH664 pKa = 6.8IGLGTNQSVDD674 pKa = 4.61LEE676 pKa = 4.55VHH678 pKa = 5.4WPSGIVEE685 pKa = 4.39TFANVAADD693 pKa = 3.49QVVEE697 pKa = 4.17LVEE700 pKa = 4.47GTGGTGPSYY709 pKa = 10.86SLSVDD714 pKa = 3.52DD715 pKa = 4.16PTVTEE720 pKa = 4.73GSDD723 pKa = 3.32TQALFTVNLSPAPGAGEE740 pKa = 4.2TVLVDD745 pKa = 3.38YY746 pKa = 8.5QTANGSAQAGSDD758 pKa = 3.68YY759 pKa = 10.85TSTSGQLSFGEE770 pKa = 4.64GDD772 pKa = 3.21SSLTVSVPILDD783 pKa = 5.22DD784 pKa = 3.55GTAEE788 pKa = 4.05PNEE791 pKa = 4.39TFTLQASSSEE801 pKa = 4.0ASPASGTATIQDD813 pKa = 4.0DD814 pKa = 4.48DD815 pKa = 5.15GSGGDD820 pKa = 4.22PTCGMPTFDD829 pKa = 3.99PAVDD833 pKa = 3.07RR834 pKa = 11.84GVYY837 pKa = 9.53LWNDD841 pKa = 3.25CGTNDD846 pKa = 2.56WHH848 pKa = 8.04LRR850 pKa = 11.84ITGGGSGQYY859 pKa = 10.34LNHH862 pKa = 7.12LGTLVADD869 pKa = 4.01APFVSVDD876 pKa = 3.45GFSVEE881 pKa = 5.15ANDD884 pKa = 4.01TLDD887 pKa = 3.53VAGSQIDD894 pKa = 3.33FGLIVGGVYY903 pKa = 10.46EE904 pKa = 4.88DD905 pKa = 5.46GIDD908 pKa = 3.54FTLAEE913 pKa = 4.42GASVCFTLTSPSGLAVFGGVDD934 pKa = 4.11ADD936 pKa = 3.91PVSLPVSMPGFGDD949 pKa = 3.64CGGGGGDD956 pKa = 4.26PTDD959 pKa = 4.24PTCGMPTFDD968 pKa = 4.21PASDD972 pKa = 3.05RR973 pKa = 11.84GVYY976 pKa = 8.89LWKK979 pKa = 10.69DD980 pKa = 3.27CGTDD984 pKa = 3.29DD985 pKa = 2.8WHH987 pKa = 8.33LRR989 pKa = 11.84ITGGSSGQYY998 pKa = 9.87LNHH1001 pKa = 6.94IGQLVADD1008 pKa = 4.53APFLDD1013 pKa = 3.76VSGVSIEE1020 pKa = 4.62GNDD1023 pKa = 3.68TLDD1026 pKa = 3.31IGASQVDD1033 pKa = 4.13FGLIVGGIYY1042 pKa = 10.25EE1043 pKa = 4.64DD1044 pKa = 5.54GIDD1047 pKa = 3.58FTLAEE1052 pKa = 4.42GASVCFSLTSPSGLSVFGGVDD1073 pKa = 3.81ADD1075 pKa = 3.96PVTLPVSMPGFGACDD1090 pKa = 3.54GGGGDD1095 pKa = 4.51PTDD1098 pKa = 4.23PTCGMPSFDD1107 pKa = 3.85PASDD1111 pKa = 3.07RR1112 pKa = 11.84GAFLWKK1118 pKa = 10.47DD1119 pKa = 3.33CGTDD1123 pKa = 3.29DD1124 pKa = 2.8WHH1126 pKa = 8.39LRR1128 pKa = 11.84ITGGGNGQYY1137 pKa = 9.88VGHH1140 pKa = 6.53VGSLVADD1147 pKa = 3.83APFVSVDD1154 pKa = 3.42GVSIEE1159 pKa = 4.65ANDD1162 pKa = 3.93TFDD1165 pKa = 3.17ITASQVDD1172 pKa = 4.11FGLIVGGIYY1181 pKa = 10.25EE1182 pKa = 4.64DD1183 pKa = 5.49GIDD1186 pKa = 3.73FTLSQGSSACFTLDD1200 pKa = 2.89SPTDD1204 pKa = 3.37VSVFGGVDD1212 pKa = 3.36GTDD1215 pKa = 2.88VGSPVGVPGFGACAVPP1231 pKa = 4.3
MM1 pKa = 7.75DD2 pKa = 5.01RR3 pKa = 11.84SRR5 pKa = 11.84FLSRR9 pKa = 11.84VGGGCLLAIGLAGAATPATAEE30 pKa = 3.83ISFSNVTGSSGITDD44 pKa = 3.38TGEE47 pKa = 4.03TFSTGWGDD55 pKa = 3.41FAGTGFPGIWLNKK68 pKa = 8.32HH69 pKa = 5.54QYY71 pKa = 7.86TPTRR75 pKa = 11.84IFYY78 pKa = 9.68NLSGSGFTEE87 pKa = 4.17VADD90 pKa = 3.92DD91 pKa = 3.67VLKK94 pKa = 11.23NEE96 pKa = 4.64EE97 pKa = 4.75GVPSPQTDD105 pKa = 3.86FNIPRR110 pKa = 11.84AEE112 pKa = 4.08TGPGEE117 pKa = 4.2FSYY120 pKa = 10.8FADD123 pKa = 3.37THH125 pKa = 6.05GQTSADD131 pKa = 3.45WDD133 pKa = 3.72NDD135 pKa = 2.85GDD137 pKa = 4.02QDD139 pKa = 4.02VLEE142 pKa = 4.38VTGAGYY148 pKa = 10.16IFPLWEE154 pKa = 4.05NDD156 pKa = 3.08GLGNFEE162 pKa = 4.09YY163 pKa = 10.61RR164 pKa = 11.84GDD166 pKa = 4.03EE167 pKa = 4.22LGFAYY172 pKa = 9.89PLTWEE177 pKa = 4.0RR178 pKa = 11.84RR179 pKa = 11.84EE180 pKa = 3.87RR181 pKa = 11.84PPIGGRR187 pKa = 11.84DD188 pKa = 3.54SLFFDD193 pKa = 4.16YY194 pKa = 11.05DD195 pKa = 3.55RR196 pKa = 11.84DD197 pKa = 3.56GWLDD201 pKa = 3.44AMVMARR207 pKa = 11.84EE208 pKa = 4.2NYY210 pKa = 8.86SAYY213 pKa = 8.47RR214 pKa = 11.84TPTAIFRR221 pKa = 11.84QEE223 pKa = 4.09RR224 pKa = 11.84IGGEE228 pKa = 4.1SFFTLDD234 pKa = 3.13EE235 pKa = 4.45STGVVPDD242 pKa = 4.12EE243 pKa = 4.46GTGPFPGTACEE254 pKa = 4.19FGAMAEE260 pKa = 4.06LSGDD264 pKa = 3.59SVLDD268 pKa = 4.17VICSDD273 pKa = 3.91SQISAIWDD281 pKa = 3.43VSQLPFADD289 pKa = 3.75LRR291 pKa = 11.84PVVGDD296 pKa = 3.92EE297 pKa = 3.72IFNRR301 pKa = 11.84YY302 pKa = 8.14PADD305 pKa = 3.53LAIGDD310 pKa = 4.31FNNDD314 pKa = 2.62LRR316 pKa = 11.84TDD318 pKa = 3.28IMAPTFAGADD328 pKa = 3.5YY329 pKa = 10.74VRR331 pKa = 11.84NLVGVVDD338 pKa = 4.62GQTLHH343 pKa = 6.07GWFNTSVPSNSGFNFSATGNVTFEE367 pKa = 4.18FDD369 pKa = 2.85WWTEE373 pKa = 3.84TPNIFIGADD382 pKa = 3.31GSNPDD387 pKa = 4.16LATNLPTMPFQEE399 pKa = 5.11LGHH402 pKa = 6.88GDD404 pKa = 3.78PNHH407 pKa = 5.9VVVTLSPTEE416 pKa = 4.08AQGLAPIFVPEE427 pKa = 5.18DD428 pKa = 3.46NDD430 pKa = 3.65GDD432 pKa = 4.83PITGVFIGYY441 pKa = 9.83VDD443 pKa = 3.81GEE445 pKa = 4.1WRR447 pKa = 11.84VRR449 pKa = 11.84FSGIEE454 pKa = 3.56RR455 pKa = 11.84WNVGMVITAQSGISGLTSVGGTKK478 pKa = 9.57IGQDD482 pKa = 3.07TGGNALLFLQSEE494 pKa = 4.32DD495 pKa = 3.76HH496 pKa = 6.62EE497 pKa = 4.96LVFSGGNVQAVNRR510 pKa = 11.84SCTAAAAADD519 pKa = 4.06FDD521 pKa = 4.55NDD523 pKa = 2.84MDD525 pKa = 5.34LDD527 pKa = 4.03VFFACSGQLSNLPDD541 pKa = 3.09IVYY544 pKa = 9.9EE545 pKa = 4.07NQGNGTFVALPAAGGAEE562 pKa = 3.96GQMPLGRR569 pKa = 11.84ADD571 pKa = 3.48SVMVADD577 pKa = 4.02YY578 pKa = 11.55DD579 pKa = 3.9EE580 pKa = 4.71DD581 pKa = 4.0CRR583 pKa = 11.84MDD585 pKa = 3.61VYY587 pKa = 10.09VTNGHH592 pKa = 6.1HH593 pKa = 6.29FRR595 pKa = 11.84PSSYY599 pKa = 11.24AGIHH603 pKa = 4.18QLFRR607 pKa = 11.84NTTSDD612 pKa = 3.58GNHH615 pKa = 7.35CIQIDD620 pKa = 4.01LEE622 pKa = 4.64GTVSNRR628 pKa = 11.84DD629 pKa = 3.31GIGAIVYY636 pKa = 7.54ATTPDD641 pKa = 3.19GTVQKK646 pKa = 10.36RR647 pKa = 11.84DD648 pKa = 3.55QNGGKK653 pKa = 9.2HH654 pKa = 6.14KK655 pKa = 10.44KK656 pKa = 8.6AQDD659 pKa = 3.54YY660 pKa = 10.16QRR662 pKa = 11.84VHH664 pKa = 6.8IGLGTNQSVDD674 pKa = 4.61LEE676 pKa = 4.55VHH678 pKa = 5.4WPSGIVEE685 pKa = 4.39TFANVAADD693 pKa = 3.49QVVEE697 pKa = 4.17LVEE700 pKa = 4.47GTGGTGPSYY709 pKa = 10.86SLSVDD714 pKa = 3.52DD715 pKa = 4.16PTVTEE720 pKa = 4.73GSDD723 pKa = 3.32TQALFTVNLSPAPGAGEE740 pKa = 4.2TVLVDD745 pKa = 3.38YY746 pKa = 8.5QTANGSAQAGSDD758 pKa = 3.68YY759 pKa = 10.85TSTSGQLSFGEE770 pKa = 4.64GDD772 pKa = 3.21SSLTVSVPILDD783 pKa = 5.22DD784 pKa = 3.55GTAEE788 pKa = 4.05PNEE791 pKa = 4.39TFTLQASSSEE801 pKa = 4.0ASPASGTATIQDD813 pKa = 4.0DD814 pKa = 4.48DD815 pKa = 5.15GSGGDD820 pKa = 4.22PTCGMPTFDD829 pKa = 3.99PAVDD833 pKa = 3.07RR834 pKa = 11.84GVYY837 pKa = 9.53LWNDD841 pKa = 3.25CGTNDD846 pKa = 2.56WHH848 pKa = 8.04LRR850 pKa = 11.84ITGGGSGQYY859 pKa = 10.34LNHH862 pKa = 7.12LGTLVADD869 pKa = 4.01APFVSVDD876 pKa = 3.45GFSVEE881 pKa = 5.15ANDD884 pKa = 4.01TLDD887 pKa = 3.53VAGSQIDD894 pKa = 3.33FGLIVGGVYY903 pKa = 10.46EE904 pKa = 4.88DD905 pKa = 5.46GIDD908 pKa = 3.54FTLAEE913 pKa = 4.42GASVCFTLTSPSGLAVFGGVDD934 pKa = 4.11ADD936 pKa = 3.91PVSLPVSMPGFGDD949 pKa = 3.64CGGGGGDD956 pKa = 4.26PTDD959 pKa = 4.24PTCGMPTFDD968 pKa = 4.21PASDD972 pKa = 3.05RR973 pKa = 11.84GVYY976 pKa = 8.89LWKK979 pKa = 10.69DD980 pKa = 3.27CGTDD984 pKa = 3.29DD985 pKa = 2.8WHH987 pKa = 8.33LRR989 pKa = 11.84ITGGSSGQYY998 pKa = 9.87LNHH1001 pKa = 6.94IGQLVADD1008 pKa = 4.53APFLDD1013 pKa = 3.76VSGVSIEE1020 pKa = 4.62GNDD1023 pKa = 3.68TLDD1026 pKa = 3.31IGASQVDD1033 pKa = 4.13FGLIVGGIYY1042 pKa = 10.25EE1043 pKa = 4.64DD1044 pKa = 5.54GIDD1047 pKa = 3.58FTLAEE1052 pKa = 4.42GASVCFSLTSPSGLSVFGGVDD1073 pKa = 3.81ADD1075 pKa = 3.96PVTLPVSMPGFGACDD1090 pKa = 3.54GGGGDD1095 pKa = 4.51PTDD1098 pKa = 4.23PTCGMPSFDD1107 pKa = 3.85PASDD1111 pKa = 3.07RR1112 pKa = 11.84GAFLWKK1118 pKa = 10.47DD1119 pKa = 3.33CGTDD1123 pKa = 3.29DD1124 pKa = 2.8WHH1126 pKa = 8.39LRR1128 pKa = 11.84ITGGGNGQYY1137 pKa = 9.88VGHH1140 pKa = 6.53VGSLVADD1147 pKa = 3.83APFVSVDD1154 pKa = 3.42GVSIEE1159 pKa = 4.65ANDD1162 pKa = 3.93TFDD1165 pKa = 3.17ITASQVDD1172 pKa = 4.11FGLIVGGIYY1181 pKa = 10.25EE1182 pKa = 4.64DD1183 pKa = 5.49GIDD1186 pKa = 3.73FTLSQGSSACFTLDD1200 pKa = 2.89SPTDD1204 pKa = 3.37VSVFGGVDD1212 pKa = 3.36GTDD1215 pKa = 2.88VGSPVGVPGFGACAVPP1231 pKa = 4.3
Molecular weight: 128.86 kDa
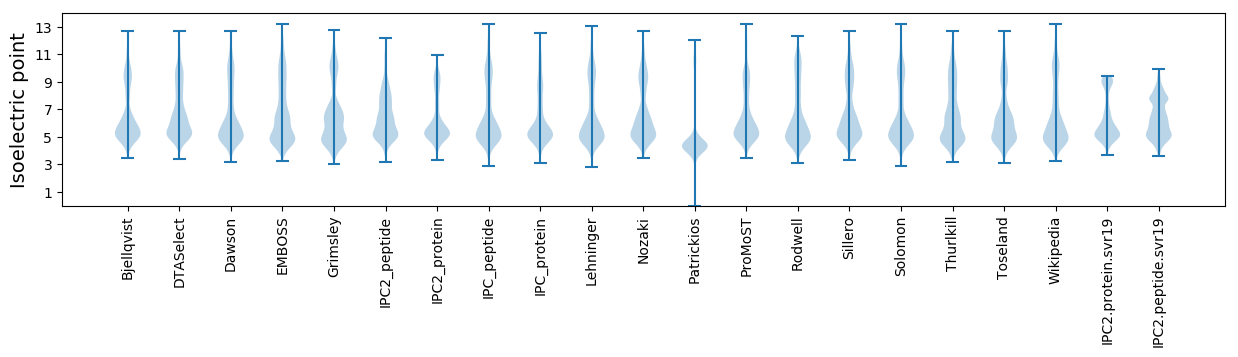
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M0K148|A0A6M0K148_9GAMM Peptide methionine sulfoxide reductase MsrA OS=Thiorhodococcus minor OX=57489 GN=msrA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LKK11 pKa = 10.48RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 6.41GFRR19 pKa = 11.84ARR21 pKa = 11.84CATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 9.43VIAARR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.62GRR39 pKa = 11.84ARR41 pKa = 11.84LAPP44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LKK11 pKa = 10.48RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 6.41GFRR19 pKa = 11.84ARR21 pKa = 11.84CATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 9.43VIAARR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.62GRR39 pKa = 11.84ARR41 pKa = 11.84LAPP44 pKa = 4.03
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1678515 |
23 |
3182 |
323.2 |
35.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.569 ± 0.039 | 1.041 ± 0.014 |
5.938 ± 0.027 | 6.428 ± 0.033 |
3.309 ± 0.018 | 8.081 ± 0.033 |
2.206 ± 0.018 | 4.733 ± 0.025 |
2.781 ± 0.027 | 11.43 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.185 ± 0.016 | 2.18 ± 0.017 |
5.278 ± 0.025 | 3.807 ± 0.022 |
7.914 ± 0.042 | 5.532 ± 0.026 |
4.859 ± 0.025 | 6.909 ± 0.033 |
1.451 ± 0.016 | 2.37 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
