
Rivularia sp. PCC 7116
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Rivulariaceae; Rivularia; unclassified Rivularia (in: Bacteria)
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
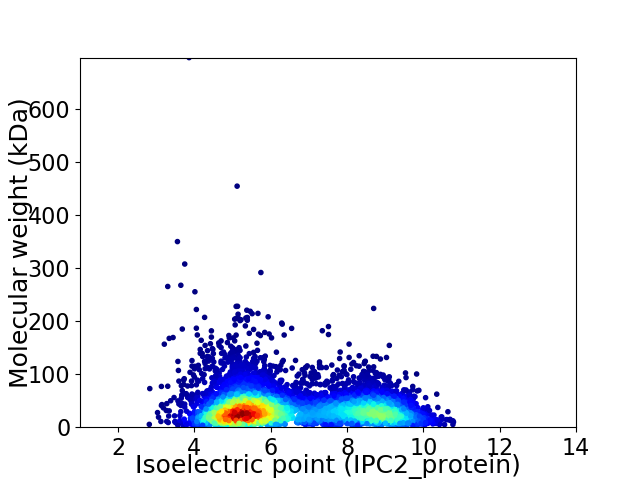
Virtual 2D-PAGE plot for 6617 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9R7V8|K9R7V8_9CYAN Uncharacterized protein OS=Rivularia sp. PCC 7116 OX=373994 GN=Riv7116_0961 PE=4 SV=1
MM1 pKa = 7.1QVNFSNSDD9 pKa = 3.61LFSEE13 pKa = 4.68SANINYY19 pKa = 7.32ITPTNEE25 pKa = 3.89LSDD28 pKa = 4.74DD29 pKa = 3.84LTSNSLNFLARR40 pKa = 11.84SSIEE44 pKa = 3.78YY45 pKa = 9.79SHH47 pKa = 6.97NDD49 pKa = 3.4SAILNQNNSDD59 pKa = 3.88LQDD62 pKa = 3.44VGSTNQLSADD72 pKa = 4.1SYY74 pKa = 11.57SSNQTGTNQTANFSALDD91 pKa = 3.46SDD93 pKa = 4.69EE94 pKa = 4.54YY95 pKa = 11.41SSEE98 pKa = 4.0TGYY101 pKa = 11.81GLVDD105 pKa = 3.62ASEE108 pKa = 4.03AVARR112 pKa = 11.84VIGEE116 pKa = 4.12DD117 pKa = 3.47TFDD120 pKa = 4.41DD121 pKa = 4.24VPDD124 pKa = 4.03KK125 pKa = 11.38GGKK128 pKa = 8.26NWGADD133 pKa = 2.89AVNAPEE139 pKa = 4.16VWEE142 pKa = 3.78QGYY145 pKa = 7.91TGEE148 pKa = 4.35GVVVAVLDD156 pKa = 3.59TGVDD160 pKa = 3.7YY161 pKa = 9.97THH163 pKa = 7.89DD164 pKa = 3.92DD165 pKa = 3.69LKK167 pKa = 11.66NNIWTNSGEE176 pKa = 4.05IEE178 pKa = 4.54NNGKK182 pKa = 9.95DD183 pKa = 3.78DD184 pKa = 5.19DD185 pKa = 4.82GNGYY189 pKa = 9.67IDD191 pKa = 6.32DD192 pKa = 4.47FYY194 pKa = 11.72GWNFDD199 pKa = 3.52GDD201 pKa = 4.14NNSTIDD207 pKa = 3.46VDD209 pKa = 3.68GHH211 pKa = 5.11GTHH214 pKa = 6.47VSGTIAGEE222 pKa = 3.99KK223 pKa = 10.25NGFGVTGIAYY233 pKa = 8.1DD234 pKa = 3.84AQIMPVKK241 pKa = 10.67VLDD244 pKa = 4.31DD245 pKa = 4.24FGSGSNTAVADD256 pKa = 4.8GIYY259 pKa = 10.32YY260 pKa = 10.66AVDD263 pKa = 3.07NGADD267 pKa = 3.88VINLSLGGSFPSFGVSEE284 pKa = 5.25AIQYY288 pKa = 10.72ASEE291 pKa = 4.08QGVTVVMAAGNSGGDD306 pKa = 3.42MPLYY310 pKa = 8.33PARR313 pKa = 11.84YY314 pKa = 8.03ADD316 pKa = 3.68EE317 pKa = 4.26YY318 pKa = 10.61GIAVGAIDD326 pKa = 4.45EE327 pKa = 4.74DD328 pKa = 4.57KK329 pKa = 11.75NMASFSNQAGSEE341 pKa = 3.98EE342 pKa = 4.42LTYY345 pKa = 10.23VTAPGVDD352 pKa = 3.24IYY354 pKa = 11.25STLPDD359 pKa = 3.75NKK361 pKa = 9.76YY362 pKa = 10.06EE363 pKa = 4.37SYY365 pKa = 10.86SGTSMATPHH374 pKa = 5.65VAGVVALMLSANPSFDD390 pKa = 3.95SNLIRR395 pKa = 11.84QTLEE399 pKa = 3.73EE400 pKa = 4.05TSKK403 pKa = 11.44NEE405 pKa = 4.41ASNQPILPDD414 pKa = 3.34LTDD417 pKa = 3.88YY418 pKa = 11.11FPDD421 pKa = 5.65LIPSGDD427 pKa = 3.65SEE429 pKa = 5.15FNFDD433 pKa = 4.19FDD435 pKa = 5.72SNPIMPSFSRR445 pKa = 11.84SFDD448 pKa = 3.22SGKK451 pKa = 9.21TMVFDD456 pKa = 4.56SSMSIDD462 pKa = 3.86KK463 pKa = 10.45SAKK466 pKa = 9.48PYY468 pKa = 10.67SEE470 pKa = 4.12SQFINYY476 pKa = 7.33QQNAWKK482 pKa = 10.45NPNNRR487 pKa = 11.84LFSDD491 pKa = 5.01DD492 pKa = 3.73IEE494 pKa = 5.48SMLEE498 pKa = 3.65EE499 pKa = 5.37DD500 pKa = 4.02EE501 pKa = 4.37VLLRR505 pKa = 11.84LEE507 pKa = 4.2RR508 pKa = 5.34
MM1 pKa = 7.1QVNFSNSDD9 pKa = 3.61LFSEE13 pKa = 4.68SANINYY19 pKa = 7.32ITPTNEE25 pKa = 3.89LSDD28 pKa = 4.74DD29 pKa = 3.84LTSNSLNFLARR40 pKa = 11.84SSIEE44 pKa = 3.78YY45 pKa = 9.79SHH47 pKa = 6.97NDD49 pKa = 3.4SAILNQNNSDD59 pKa = 3.88LQDD62 pKa = 3.44VGSTNQLSADD72 pKa = 4.1SYY74 pKa = 11.57SSNQTGTNQTANFSALDD91 pKa = 3.46SDD93 pKa = 4.69EE94 pKa = 4.54YY95 pKa = 11.41SSEE98 pKa = 4.0TGYY101 pKa = 11.81GLVDD105 pKa = 3.62ASEE108 pKa = 4.03AVARR112 pKa = 11.84VIGEE116 pKa = 4.12DD117 pKa = 3.47TFDD120 pKa = 4.41DD121 pKa = 4.24VPDD124 pKa = 4.03KK125 pKa = 11.38GGKK128 pKa = 8.26NWGADD133 pKa = 2.89AVNAPEE139 pKa = 4.16VWEE142 pKa = 3.78QGYY145 pKa = 7.91TGEE148 pKa = 4.35GVVVAVLDD156 pKa = 3.59TGVDD160 pKa = 3.7YY161 pKa = 9.97THH163 pKa = 7.89DD164 pKa = 3.92DD165 pKa = 3.69LKK167 pKa = 11.66NNIWTNSGEE176 pKa = 4.05IEE178 pKa = 4.54NNGKK182 pKa = 9.95DD183 pKa = 3.78DD184 pKa = 5.19DD185 pKa = 4.82GNGYY189 pKa = 9.67IDD191 pKa = 6.32DD192 pKa = 4.47FYY194 pKa = 11.72GWNFDD199 pKa = 3.52GDD201 pKa = 4.14NNSTIDD207 pKa = 3.46VDD209 pKa = 3.68GHH211 pKa = 5.11GTHH214 pKa = 6.47VSGTIAGEE222 pKa = 3.99KK223 pKa = 10.25NGFGVTGIAYY233 pKa = 8.1DD234 pKa = 3.84AQIMPVKK241 pKa = 10.67VLDD244 pKa = 4.31DD245 pKa = 4.24FGSGSNTAVADD256 pKa = 4.8GIYY259 pKa = 10.32YY260 pKa = 10.66AVDD263 pKa = 3.07NGADD267 pKa = 3.88VINLSLGGSFPSFGVSEE284 pKa = 5.25AIQYY288 pKa = 10.72ASEE291 pKa = 4.08QGVTVVMAAGNSGGDD306 pKa = 3.42MPLYY310 pKa = 8.33PARR313 pKa = 11.84YY314 pKa = 8.03ADD316 pKa = 3.68EE317 pKa = 4.26YY318 pKa = 10.61GIAVGAIDD326 pKa = 4.45EE327 pKa = 4.74DD328 pKa = 4.57KK329 pKa = 11.75NMASFSNQAGSEE341 pKa = 3.98EE342 pKa = 4.42LTYY345 pKa = 10.23VTAPGVDD352 pKa = 3.24IYY354 pKa = 11.25STLPDD359 pKa = 3.75NKK361 pKa = 9.76YY362 pKa = 10.06EE363 pKa = 4.37SYY365 pKa = 10.86SGTSMATPHH374 pKa = 5.65VAGVVALMLSANPSFDD390 pKa = 3.95SNLIRR395 pKa = 11.84QTLEE399 pKa = 3.73EE400 pKa = 4.05TSKK403 pKa = 11.44NEE405 pKa = 4.41ASNQPILPDD414 pKa = 3.34LTDD417 pKa = 3.88YY418 pKa = 11.11FPDD421 pKa = 5.65LIPSGDD427 pKa = 3.65SEE429 pKa = 5.15FNFDD433 pKa = 4.19FDD435 pKa = 5.72SNPIMPSFSRR445 pKa = 11.84SFDD448 pKa = 3.22SGKK451 pKa = 9.21TMVFDD456 pKa = 4.56SSMSIDD462 pKa = 3.86KK463 pKa = 10.45SAKK466 pKa = 9.48PYY468 pKa = 10.67SEE470 pKa = 4.12SQFINYY476 pKa = 7.33QQNAWKK482 pKa = 10.45NPNNRR487 pKa = 11.84LFSDD491 pKa = 5.01DD492 pKa = 3.73IEE494 pKa = 5.48SMLEE498 pKa = 3.65EE499 pKa = 5.37DD500 pKa = 4.02EE501 pKa = 4.37VLLRR505 pKa = 11.84LEE507 pKa = 4.2RR508 pKa = 5.34
Molecular weight: 54.75 kDa
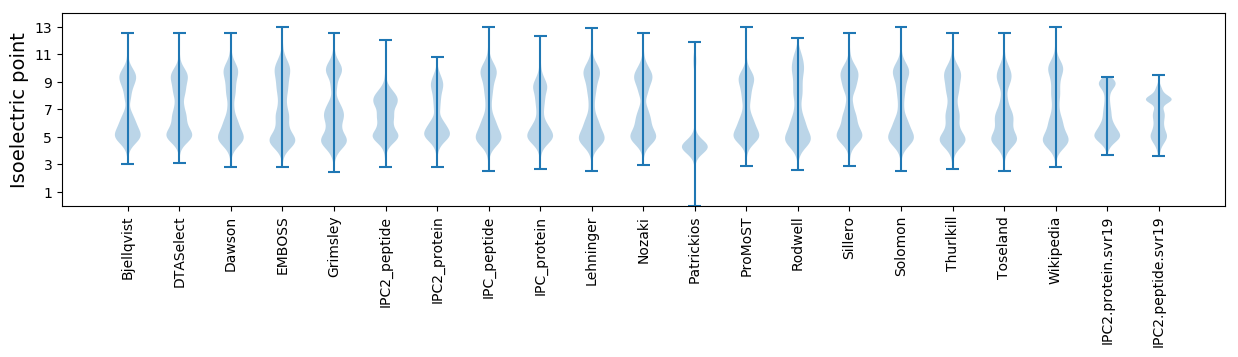
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9R5Q9|K9R5Q9_9CYAN Uncharacterized protein OS=Rivularia sp. PCC 7116 OX=373994 GN=Riv7116_0077 PE=4 SV=1
MM1 pKa = 7.39KK2 pKa = 8.93RR3 pKa = 11.84TLGGTCRR10 pKa = 11.84KK11 pKa = 9.61RR12 pKa = 11.84KK13 pKa = 7.61RR14 pKa = 11.84TSGFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TPDD26 pKa = 3.17GRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.01KK37 pKa = 9.6GRR39 pKa = 11.84HH40 pKa = 5.0RR41 pKa = 11.84LSVV44 pKa = 3.12
MM1 pKa = 7.39KK2 pKa = 8.93RR3 pKa = 11.84TLGGTCRR10 pKa = 11.84KK11 pKa = 9.61RR12 pKa = 11.84KK13 pKa = 7.61RR14 pKa = 11.84TSGFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TPDD26 pKa = 3.17GRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.01KK37 pKa = 9.6GRR39 pKa = 11.84HH40 pKa = 5.0RR41 pKa = 11.84LSVV44 pKa = 3.12
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2262525 |
29 |
6379 |
341.9 |
38.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.54 ± 0.033 | 0.968 ± 0.011 |
5.013 ± 0.031 | 6.543 ± 0.035 |
4.193 ± 0.026 | 6.546 ± 0.037 |
1.601 ± 0.017 | 7.251 ± 0.028 |
5.836 ± 0.035 | 10.469 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.785 ± 0.015 | 5.348 ± 0.037 |
4.212 ± 0.023 | 4.897 ± 0.028 |
4.661 ± 0.026 | 6.887 ± 0.026 |
5.465 ± 0.033 | 6.376 ± 0.023 |
1.315 ± 0.014 | 3.094 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
