
Hubei tombus-like virus 8
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.6
Get precalculated fractions of proteins
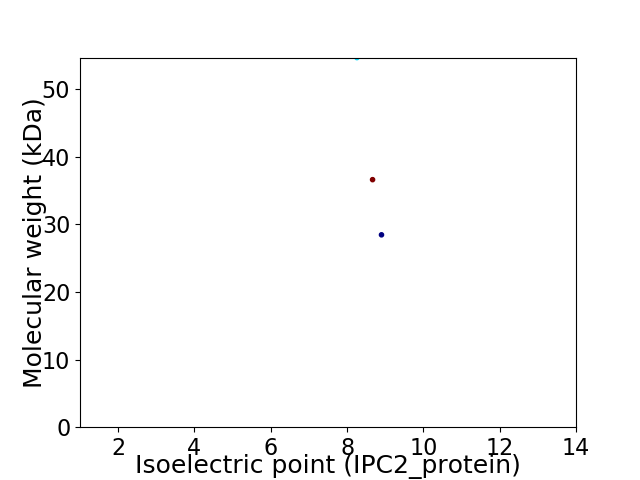
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
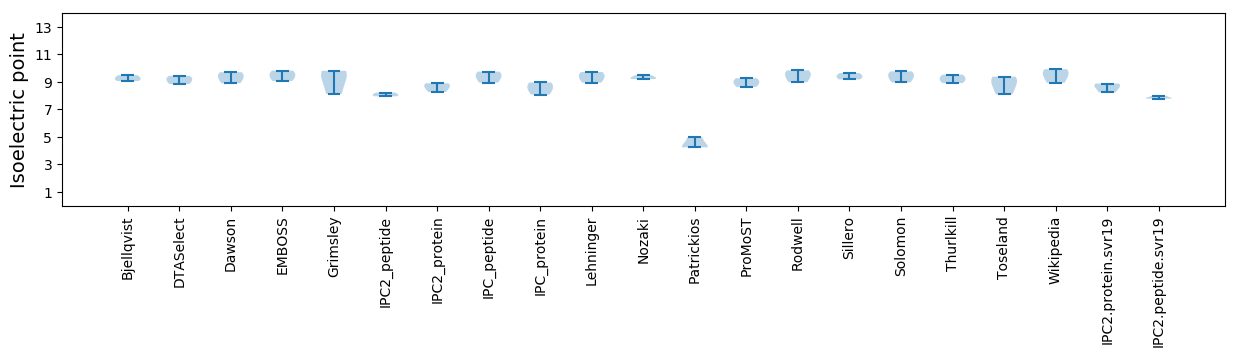
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGY5|A0A1L3KGY5_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 8 OX=1923295 PE=4 SV=1
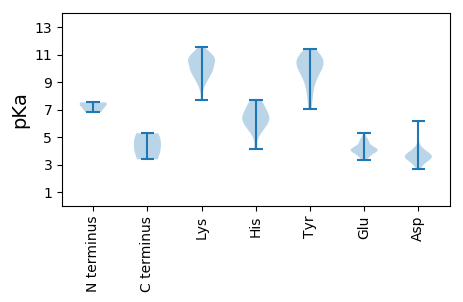
MM1 pKa = 7.52RR2 pKa = 11.84AVDD5 pKa = 3.17RR6 pKa = 11.84ALRR9 pKa = 11.84EE10 pKa = 3.63RR11 pKa = 11.84YY12 pKa = 9.52FFVKK16 pKa = 9.44TKK18 pKa = 10.88KK19 pKa = 10.42EE20 pKa = 4.12GFQPALPVRR29 pKa = 11.84SRR31 pKa = 11.84AYY33 pKa = 8.7EE34 pKa = 3.86TDD36 pKa = 2.68VHH38 pKa = 6.48LRR40 pKa = 11.84SFRR43 pKa = 11.84AQVVASCKK51 pKa = 10.55APVVSIADD59 pKa = 3.48VVNSYY64 pKa = 9.11TGPKK68 pKa = 9.36RR69 pKa = 11.84EE70 pKa = 4.59AYY72 pKa = 9.76AKK74 pKa = 10.57AAISLMKK81 pKa = 10.53DD82 pKa = 3.06AVNHH86 pKa = 5.64QDD88 pKa = 2.92ASLMMFGKK96 pKa = 10.4FEE98 pKa = 4.03KK99 pKa = 10.62QNLDD103 pKa = 3.06KK104 pKa = 11.1ANRR107 pKa = 11.84GINPRR112 pKa = 11.84SKK114 pKa = 10.52RR115 pKa = 11.84YY116 pKa = 8.82NLEE119 pKa = 3.54LGRR122 pKa = 11.84YY123 pKa = 8.07LKK125 pKa = 10.8FMEE128 pKa = 4.72KK129 pKa = 9.92PLYY132 pKa = 10.2RR133 pKa = 11.84SINKK137 pKa = 10.01AYY139 pKa = 10.21GARR142 pKa = 11.84TAHH145 pKa = 5.47TVIKK149 pKa = 9.51GLNVADD155 pKa = 4.09SAAVAHH161 pKa = 6.37AKK163 pKa = 8.94WKK165 pKa = 9.89LFKK168 pKa = 10.84RR169 pKa = 11.84PVAVGLDD176 pKa = 3.5AEE178 pKa = 4.74KK179 pKa = 10.68FDD181 pKa = 3.67AHH183 pKa = 7.66VSIPCLRR190 pKa = 11.84YY191 pKa = 8.2EE192 pKa = 3.98HH193 pKa = 6.81SFYY196 pKa = 11.16LQIHH200 pKa = 5.4SRR202 pKa = 11.84QVKK205 pKa = 8.37RR206 pKa = 11.84QMMLKK211 pKa = 9.89QLLNWQLVNTGVAYY225 pKa = 10.87CDD227 pKa = 3.36DD228 pKa = 3.92GKK230 pKa = 11.02IKK232 pKa = 10.64FKK234 pKa = 10.89IKK236 pKa = 9.14GTRR239 pKa = 11.84CSGDD243 pKa = 3.56LNTSMGNCLIMCGLVYY259 pKa = 10.26AYY261 pKa = 9.75SASMGVNVEE270 pKa = 4.08LMNNGDD276 pKa = 3.85DD277 pKa = 3.65CVVIMEE283 pKa = 4.49EE284 pKa = 4.09EE285 pKa = 4.06DD286 pKa = 3.51VEE288 pKa = 4.92KK289 pKa = 11.07YY290 pKa = 9.8VAGVPEE296 pKa = 4.01YY297 pKa = 10.48FKK299 pKa = 11.11RR300 pKa = 11.84KK301 pKa = 7.68GFRR304 pKa = 11.84MTVEE308 pKa = 3.82EE309 pKa = 4.17PVRR312 pKa = 11.84EE313 pKa = 3.93FEE315 pKa = 5.1QIEE318 pKa = 4.74FCQSHH323 pKa = 6.52PVHH326 pKa = 7.09DD327 pKa = 4.16GEE329 pKa = 4.78KK330 pKa = 9.28YY331 pKa = 10.19VMVRR335 pKa = 11.84NLTNCLQKK343 pKa = 11.17DD344 pKa = 4.0PMCLVPIQNAKK355 pKa = 10.38ALQQWYY361 pKa = 9.09DD362 pKa = 3.43AVGSCGLSLTPGIPVLQEE380 pKa = 3.61FYY382 pKa = 10.0KK383 pKa = 9.73TFKK386 pKa = 10.66RR387 pKa = 11.84SGRR390 pKa = 11.84PCRR393 pKa = 11.84EE394 pKa = 3.52GFKK397 pKa = 10.17RR398 pKa = 11.84TVFKK402 pKa = 10.13NTSYY406 pKa = 11.03YY407 pKa = 10.71EE408 pKa = 3.97RR409 pKa = 11.84VKK411 pKa = 11.1DD412 pKa = 3.45LTGRR416 pKa = 11.84EE417 pKa = 4.28LSCSSVSRR425 pKa = 11.84SSFYY429 pKa = 10.65FAFGVLPEE437 pKa = 4.02HH438 pKa = 6.56QIAIEE443 pKa = 4.17AYY445 pKa = 7.87YY446 pKa = 11.39ASMTLSAVVEE456 pKa = 4.41DD457 pKa = 4.42AMHH460 pKa = 7.28PEE462 pKa = 3.74YY463 pKa = 10.69AYY465 pKa = 10.81DD466 pKa = 5.4KK467 pKa = 11.2GDD469 pKa = 3.55NCCPPVVDD477 pKa = 6.16CIFAA481 pKa = 4.4
MM1 pKa = 7.52RR2 pKa = 11.84AVDD5 pKa = 3.17RR6 pKa = 11.84ALRR9 pKa = 11.84EE10 pKa = 3.63RR11 pKa = 11.84YY12 pKa = 9.52FFVKK16 pKa = 9.44TKK18 pKa = 10.88KK19 pKa = 10.42EE20 pKa = 4.12GFQPALPVRR29 pKa = 11.84SRR31 pKa = 11.84AYY33 pKa = 8.7EE34 pKa = 3.86TDD36 pKa = 2.68VHH38 pKa = 6.48LRR40 pKa = 11.84SFRR43 pKa = 11.84AQVVASCKK51 pKa = 10.55APVVSIADD59 pKa = 3.48VVNSYY64 pKa = 9.11TGPKK68 pKa = 9.36RR69 pKa = 11.84EE70 pKa = 4.59AYY72 pKa = 9.76AKK74 pKa = 10.57AAISLMKK81 pKa = 10.53DD82 pKa = 3.06AVNHH86 pKa = 5.64QDD88 pKa = 2.92ASLMMFGKK96 pKa = 10.4FEE98 pKa = 4.03KK99 pKa = 10.62QNLDD103 pKa = 3.06KK104 pKa = 11.1ANRR107 pKa = 11.84GINPRR112 pKa = 11.84SKK114 pKa = 10.52RR115 pKa = 11.84YY116 pKa = 8.82NLEE119 pKa = 3.54LGRR122 pKa = 11.84YY123 pKa = 8.07LKK125 pKa = 10.8FMEE128 pKa = 4.72KK129 pKa = 9.92PLYY132 pKa = 10.2RR133 pKa = 11.84SINKK137 pKa = 10.01AYY139 pKa = 10.21GARR142 pKa = 11.84TAHH145 pKa = 5.47TVIKK149 pKa = 9.51GLNVADD155 pKa = 4.09SAAVAHH161 pKa = 6.37AKK163 pKa = 8.94WKK165 pKa = 9.89LFKK168 pKa = 10.84RR169 pKa = 11.84PVAVGLDD176 pKa = 3.5AEE178 pKa = 4.74KK179 pKa = 10.68FDD181 pKa = 3.67AHH183 pKa = 7.66VSIPCLRR190 pKa = 11.84YY191 pKa = 8.2EE192 pKa = 3.98HH193 pKa = 6.81SFYY196 pKa = 11.16LQIHH200 pKa = 5.4SRR202 pKa = 11.84QVKK205 pKa = 8.37RR206 pKa = 11.84QMMLKK211 pKa = 9.89QLLNWQLVNTGVAYY225 pKa = 10.87CDD227 pKa = 3.36DD228 pKa = 3.92GKK230 pKa = 11.02IKK232 pKa = 10.64FKK234 pKa = 10.89IKK236 pKa = 9.14GTRR239 pKa = 11.84CSGDD243 pKa = 3.56LNTSMGNCLIMCGLVYY259 pKa = 10.26AYY261 pKa = 9.75SASMGVNVEE270 pKa = 4.08LMNNGDD276 pKa = 3.85DD277 pKa = 3.65CVVIMEE283 pKa = 4.49EE284 pKa = 4.09EE285 pKa = 4.06DD286 pKa = 3.51VEE288 pKa = 4.92KK289 pKa = 11.07YY290 pKa = 9.8VAGVPEE296 pKa = 4.01YY297 pKa = 10.48FKK299 pKa = 11.11RR300 pKa = 11.84KK301 pKa = 7.68GFRR304 pKa = 11.84MTVEE308 pKa = 3.82EE309 pKa = 4.17PVRR312 pKa = 11.84EE313 pKa = 3.93FEE315 pKa = 5.1QIEE318 pKa = 4.74FCQSHH323 pKa = 6.52PVHH326 pKa = 7.09DD327 pKa = 4.16GEE329 pKa = 4.78KK330 pKa = 9.28YY331 pKa = 10.19VMVRR335 pKa = 11.84NLTNCLQKK343 pKa = 11.17DD344 pKa = 4.0PMCLVPIQNAKK355 pKa = 10.38ALQQWYY361 pKa = 9.09DD362 pKa = 3.43AVGSCGLSLTPGIPVLQEE380 pKa = 3.61FYY382 pKa = 10.0KK383 pKa = 9.73TFKK386 pKa = 10.66RR387 pKa = 11.84SGRR390 pKa = 11.84PCRR393 pKa = 11.84EE394 pKa = 3.52GFKK397 pKa = 10.17RR398 pKa = 11.84TVFKK402 pKa = 10.13NTSYY406 pKa = 11.03YY407 pKa = 10.71EE408 pKa = 3.97RR409 pKa = 11.84VKK411 pKa = 11.1DD412 pKa = 3.45LTGRR416 pKa = 11.84EE417 pKa = 4.28LSCSSVSRR425 pKa = 11.84SSFYY429 pKa = 10.65FAFGVLPEE437 pKa = 4.02HH438 pKa = 6.56QIAIEE443 pKa = 4.17AYY445 pKa = 7.87YY446 pKa = 11.39ASMTLSAVVEE456 pKa = 4.41DD457 pKa = 4.42AMHH460 pKa = 7.28PEE462 pKa = 3.74YY463 pKa = 10.69AYY465 pKa = 10.81DD466 pKa = 5.4KK467 pKa = 11.2GDD469 pKa = 3.55NCCPPVVDD477 pKa = 6.16CIFAA481 pKa = 4.4
Molecular weight: 54.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGY5|A0A1L3KGY5_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 8 OX=1923295 PE=4 SV=1
MM1 pKa = 7.39QNNFYY6 pKa = 10.45EE7 pKa = 4.33KK8 pKa = 10.69RR9 pKa = 11.84EE10 pKa = 3.9FQVNEE15 pKa = 3.34VSARR19 pKa = 11.84HH20 pKa = 4.13TRR22 pKa = 11.84EE23 pKa = 3.63RR24 pKa = 11.84RR25 pKa = 11.84NLWAEE30 pKa = 3.57KK31 pKa = 9.27WDD33 pKa = 3.78SFKK36 pKa = 11.44SGFAEE41 pKa = 4.18MYY43 pKa = 7.09YY44 pKa = 10.45TSTFFNLLCCTVPEE58 pKa = 4.12DD59 pKa = 3.77HH60 pKa = 7.53LEE62 pKa = 4.26FKK64 pKa = 10.77LDD66 pKa = 3.46QADD69 pKa = 3.79RR70 pKa = 11.84LAVRR74 pKa = 11.84KK75 pKa = 9.82ALMQSLDD82 pKa = 3.61YY83 pKa = 10.46GAHH86 pKa = 6.36KK87 pKa = 10.51SIIAATMEE95 pKa = 5.31DD96 pKa = 3.39ISEE99 pKa = 4.31HH100 pKa = 5.97EE101 pKa = 4.64GYY103 pKa = 10.2EE104 pKa = 3.92MRR106 pKa = 11.84MDD108 pKa = 4.03PPVKK112 pKa = 9.75QVPVPSSSSTALIVRR127 pKa = 11.84NVNLQPQGAVMLKK140 pKa = 9.83PVKK143 pKa = 9.99GVQGVRR149 pKa = 11.84SIPRR153 pKa = 11.84FTAAVVVALRR163 pKa = 11.84GKK165 pKa = 10.21LGQLNPSVQGNEE177 pKa = 3.71LVLEE181 pKa = 4.34RR182 pKa = 11.84EE183 pKa = 4.89ALRR186 pKa = 11.84MMRR189 pKa = 11.84DD190 pKa = 3.46YY191 pKa = 11.23NVRR194 pKa = 11.84EE195 pKa = 3.98VDD197 pKa = 3.74RR198 pKa = 11.84AVHH201 pKa = 6.35LPRR204 pKa = 11.84VMAAYY209 pKa = 9.15WSADD213 pKa = 2.9VHH215 pKa = 6.03YY216 pKa = 9.89RR217 pKa = 11.84VPTVEE222 pKa = 3.94SRR224 pKa = 11.84MSRR227 pKa = 11.84FQRR230 pKa = 11.84WLLNRR235 pKa = 11.84PSSSPQYY242 pKa = 10.53AVPRR246 pKa = 11.84AA247 pKa = 3.38
MM1 pKa = 7.39QNNFYY6 pKa = 10.45EE7 pKa = 4.33KK8 pKa = 10.69RR9 pKa = 11.84EE10 pKa = 3.9FQVNEE15 pKa = 3.34VSARR19 pKa = 11.84HH20 pKa = 4.13TRR22 pKa = 11.84EE23 pKa = 3.63RR24 pKa = 11.84RR25 pKa = 11.84NLWAEE30 pKa = 3.57KK31 pKa = 9.27WDD33 pKa = 3.78SFKK36 pKa = 11.44SGFAEE41 pKa = 4.18MYY43 pKa = 7.09YY44 pKa = 10.45TSTFFNLLCCTVPEE58 pKa = 4.12DD59 pKa = 3.77HH60 pKa = 7.53LEE62 pKa = 4.26FKK64 pKa = 10.77LDD66 pKa = 3.46QADD69 pKa = 3.79RR70 pKa = 11.84LAVRR74 pKa = 11.84KK75 pKa = 9.82ALMQSLDD82 pKa = 3.61YY83 pKa = 10.46GAHH86 pKa = 6.36KK87 pKa = 10.51SIIAATMEE95 pKa = 5.31DD96 pKa = 3.39ISEE99 pKa = 4.31HH100 pKa = 5.97EE101 pKa = 4.64GYY103 pKa = 10.2EE104 pKa = 3.92MRR106 pKa = 11.84MDD108 pKa = 4.03PPVKK112 pKa = 9.75QVPVPSSSSTALIVRR127 pKa = 11.84NVNLQPQGAVMLKK140 pKa = 9.83PVKK143 pKa = 9.99GVQGVRR149 pKa = 11.84SIPRR153 pKa = 11.84FTAAVVVALRR163 pKa = 11.84GKK165 pKa = 10.21LGQLNPSVQGNEE177 pKa = 3.71LVLEE181 pKa = 4.34RR182 pKa = 11.84EE183 pKa = 4.89ALRR186 pKa = 11.84MMRR189 pKa = 11.84DD190 pKa = 3.46YY191 pKa = 11.23NVRR194 pKa = 11.84EE195 pKa = 3.98VDD197 pKa = 3.74RR198 pKa = 11.84AVHH201 pKa = 6.35LPRR204 pKa = 11.84VMAAYY209 pKa = 9.15WSADD213 pKa = 2.9VHH215 pKa = 6.03YY216 pKa = 9.89RR217 pKa = 11.84VPTVEE222 pKa = 3.94SRR224 pKa = 11.84MSRR227 pKa = 11.84FQRR230 pKa = 11.84WLLNRR235 pKa = 11.84PSSSPQYY242 pKa = 10.53AVPRR246 pKa = 11.84AA247 pKa = 3.38
Molecular weight: 28.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1070 |
247 |
481 |
356.7 |
39.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.159 ± 0.492 | 2.15 ± 0.63 |
4.112 ± 0.353 | 5.327 ± 0.801 |
4.393 ± 0.271 | 6.168 ± 1.081 |
1.776 ± 0.432 | 3.458 ± 0.511 |
5.888 ± 0.939 | 7.196 ± 0.309 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.364 ± 0.444 | 4.953 ± 0.53 |
4.86 ± 0.377 | 3.645 ± 0.425 |
6.542 ± 1.144 | 6.822 ± 0.348 |
5.888 ± 1.984 | 8.879 ± 0.676 |
0.841 ± 0.262 | 4.579 ± 0.371 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
