
Nostoc sp. PCC 7107
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Nostocaceae; Nostoc; unclassified Nostoc
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
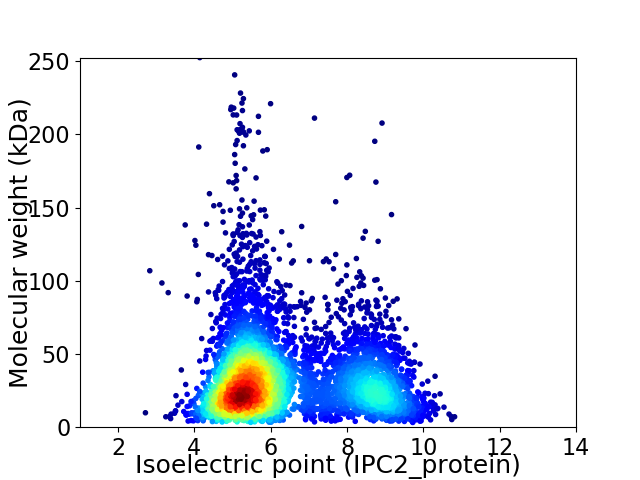
Virtual 2D-PAGE plot for 5185 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9QBW6|K9QBW6_9NOSO Histidine kinase OS=Nostoc sp. PCC 7107 OX=317936 GN=Nos7107_2027 PE=4 SV=1
MM1 pKa = 6.68TTIPSDD7 pKa = 3.72PLFPNQWYY15 pKa = 9.91LYY17 pKa = 9.78NQGQSGGTPGIDD29 pKa = 2.91LNVVNVWEE37 pKa = 5.15DD38 pKa = 3.35YY39 pKa = 8.86TGRR42 pKa = 11.84GVVVGVIDD50 pKa = 4.99DD51 pKa = 4.25GFDD54 pKa = 3.34YY55 pKa = 10.59THH57 pKa = 7.09YY58 pKa = 11.3DD59 pKa = 3.33LDD61 pKa = 4.04NNYY64 pKa = 7.99DD65 pKa = 3.43TTRR68 pKa = 11.84DD69 pKa = 3.42YY70 pKa = 11.31DD71 pKa = 4.0TIEE74 pKa = 4.02QNFDD78 pKa = 3.37PFGNAIDD85 pKa = 3.69SHH87 pKa = 5.71GTSVAGIIASEE98 pKa = 4.2ANGVGTVGVAFNATISGIRR117 pKa = 11.84AIGFGGDD124 pKa = 3.26VVGALWRR131 pKa = 11.84AEE133 pKa = 4.05SFDD136 pKa = 3.58VVNNSWGVSGFFADD150 pKa = 3.72NFYY153 pKa = 10.41TIPQAGQAIANAVTNGRR170 pKa = 11.84NGLGTAIVFAAGNSRR185 pKa = 11.84QEE187 pKa = 4.09GDD189 pKa = 2.95NTNYY193 pKa = 10.42HH194 pKa = 6.09NFQNSRR200 pKa = 11.84QVITVAALDD209 pKa = 4.03HH210 pKa = 6.72NGFASPYY217 pKa = 7.46STPGASILVSAFGSGYY233 pKa = 10.48DD234 pKa = 3.76GSIVTTDD241 pKa = 3.14RR242 pKa = 11.84QGSDD246 pKa = 2.96GYY248 pKa = 11.37SFEE251 pKa = 5.13DD252 pKa = 3.42YY253 pKa = 10.67NYY255 pKa = 10.78SFNGTSAAAPQVSGVVALLLEE276 pKa = 4.61ANRR279 pKa = 11.84NLGYY283 pKa = 10.3RR284 pKa = 11.84DD285 pKa = 3.26IQEE288 pKa = 3.85ILAYY292 pKa = 9.4SARR295 pKa = 11.84QNDD298 pKa = 4.75FYY300 pKa = 11.6NQGGNYY306 pKa = 8.18IWQINGANNFNGGGLHH322 pKa = 6.22VSHH325 pKa = 8.04DD326 pKa = 3.98YY327 pKa = 11.66GFGLVDD333 pKa = 3.1AHH335 pKa = 6.37AAVRR339 pKa = 11.84LAEE342 pKa = 4.16TWQKK346 pKa = 11.04QSVFSNEE353 pKa = 3.39QSLSYY358 pKa = 11.01SSGNLGLVIPDD369 pKa = 4.02SNTGGISSTFTVAAGLDD386 pKa = 3.8VDD388 pKa = 3.86WVEE391 pKa = 4.25VEE393 pKa = 4.73LNLTHH398 pKa = 7.29SYY400 pKa = 10.71RR401 pKa = 11.84GDD403 pKa = 3.57LVVDD407 pKa = 5.54LISPSGTISRR417 pKa = 11.84LINQPGDD424 pKa = 3.14GWDD427 pKa = 3.35SGDD430 pKa = 3.67NIVFKK435 pKa = 10.87LSSSQHH441 pKa = 5.28WGEE444 pKa = 4.07SSAGNWTLRR453 pKa = 11.84IQDD456 pKa = 4.71LFLSDD461 pKa = 3.64TGILNSWNLRR471 pKa = 11.84LYY473 pKa = 11.15GDD475 pKa = 3.91TDD477 pKa = 3.93TANDD481 pKa = 3.71TYY483 pKa = 10.93FYY485 pKa = 8.94TNEE488 pKa = 3.65YY489 pKa = 9.88GFYY492 pKa = 10.03GTTTPSDD499 pKa = 3.37NTGTDD504 pKa = 3.67TINAAAITSNSYY516 pKa = 11.36LDD518 pKa = 4.58LNAGSTSILNGTTLIINTGTIIEE541 pKa = 4.29NAFGGDD547 pKa = 3.62GNDD550 pKa = 3.74TIIGNSAANVLSGGRR565 pKa = 11.84GDD567 pKa = 5.05DD568 pKa = 3.99SLDD571 pKa = 3.53GGAGNDD577 pKa = 3.64TLRR580 pKa = 11.84GGRR583 pKa = 11.84DD584 pKa = 2.99NDD586 pKa = 3.92TYY588 pKa = 11.13IVNTSGDD595 pKa = 3.85VVIEE599 pKa = 3.99NANEE603 pKa = 4.59GIDD606 pKa = 3.88TIRR609 pKa = 11.84TSISYY614 pKa = 8.57TLGANLEE621 pKa = 4.04NLILTGTAAINGTGNTLNNSITGNSGNNTLNGGVGNDD658 pKa = 3.93TLNGGAGNDD667 pKa = 3.73TYY669 pKa = 11.14TVDD672 pKa = 3.67SIGDD676 pKa = 3.69VVIEE680 pKa = 4.12NANEE684 pKa = 3.94GTDD687 pKa = 3.75TIRR690 pKa = 11.84TSISYY695 pKa = 8.62TLGSNLEE702 pKa = 3.95NLILTGTAAINGTGNTLNNSITGNSGNNTLNGGAGNDD739 pKa = 3.73TLTAGAGNDD748 pKa = 3.65TLTGGLGRR756 pKa = 11.84DD757 pKa = 3.08SFNFNSRR764 pKa = 11.84TEE766 pKa = 4.46GIDD769 pKa = 3.41RR770 pKa = 11.84ITDD773 pKa = 3.8FSVIDD778 pKa = 3.5DD779 pKa = 4.61TIFVSASGFGGGLVTGAAIAANRR802 pKa = 11.84FVIGAAATTSSQRR815 pKa = 11.84FIYY818 pKa = 10.88NNVTGGLFFDD828 pKa = 4.87ADD830 pKa = 3.73GTGAIARR837 pKa = 11.84IQFATLSAGLSLTNADD853 pKa = 3.29IFVNTT858 pKa = 4.48
MM1 pKa = 6.68TTIPSDD7 pKa = 3.72PLFPNQWYY15 pKa = 9.91LYY17 pKa = 9.78NQGQSGGTPGIDD29 pKa = 2.91LNVVNVWEE37 pKa = 5.15DD38 pKa = 3.35YY39 pKa = 8.86TGRR42 pKa = 11.84GVVVGVIDD50 pKa = 4.99DD51 pKa = 4.25GFDD54 pKa = 3.34YY55 pKa = 10.59THH57 pKa = 7.09YY58 pKa = 11.3DD59 pKa = 3.33LDD61 pKa = 4.04NNYY64 pKa = 7.99DD65 pKa = 3.43TTRR68 pKa = 11.84DD69 pKa = 3.42YY70 pKa = 11.31DD71 pKa = 4.0TIEE74 pKa = 4.02QNFDD78 pKa = 3.37PFGNAIDD85 pKa = 3.69SHH87 pKa = 5.71GTSVAGIIASEE98 pKa = 4.2ANGVGTVGVAFNATISGIRR117 pKa = 11.84AIGFGGDD124 pKa = 3.26VVGALWRR131 pKa = 11.84AEE133 pKa = 4.05SFDD136 pKa = 3.58VVNNSWGVSGFFADD150 pKa = 3.72NFYY153 pKa = 10.41TIPQAGQAIANAVTNGRR170 pKa = 11.84NGLGTAIVFAAGNSRR185 pKa = 11.84QEE187 pKa = 4.09GDD189 pKa = 2.95NTNYY193 pKa = 10.42HH194 pKa = 6.09NFQNSRR200 pKa = 11.84QVITVAALDD209 pKa = 4.03HH210 pKa = 6.72NGFASPYY217 pKa = 7.46STPGASILVSAFGSGYY233 pKa = 10.48DD234 pKa = 3.76GSIVTTDD241 pKa = 3.14RR242 pKa = 11.84QGSDD246 pKa = 2.96GYY248 pKa = 11.37SFEE251 pKa = 5.13DD252 pKa = 3.42YY253 pKa = 10.67NYY255 pKa = 10.78SFNGTSAAAPQVSGVVALLLEE276 pKa = 4.61ANRR279 pKa = 11.84NLGYY283 pKa = 10.3RR284 pKa = 11.84DD285 pKa = 3.26IQEE288 pKa = 3.85ILAYY292 pKa = 9.4SARR295 pKa = 11.84QNDD298 pKa = 4.75FYY300 pKa = 11.6NQGGNYY306 pKa = 8.18IWQINGANNFNGGGLHH322 pKa = 6.22VSHH325 pKa = 8.04DD326 pKa = 3.98YY327 pKa = 11.66GFGLVDD333 pKa = 3.1AHH335 pKa = 6.37AAVRR339 pKa = 11.84LAEE342 pKa = 4.16TWQKK346 pKa = 11.04QSVFSNEE353 pKa = 3.39QSLSYY358 pKa = 11.01SSGNLGLVIPDD369 pKa = 4.02SNTGGISSTFTVAAGLDD386 pKa = 3.8VDD388 pKa = 3.86WVEE391 pKa = 4.25VEE393 pKa = 4.73LNLTHH398 pKa = 7.29SYY400 pKa = 10.71RR401 pKa = 11.84GDD403 pKa = 3.57LVVDD407 pKa = 5.54LISPSGTISRR417 pKa = 11.84LINQPGDD424 pKa = 3.14GWDD427 pKa = 3.35SGDD430 pKa = 3.67NIVFKK435 pKa = 10.87LSSSQHH441 pKa = 5.28WGEE444 pKa = 4.07SSAGNWTLRR453 pKa = 11.84IQDD456 pKa = 4.71LFLSDD461 pKa = 3.64TGILNSWNLRR471 pKa = 11.84LYY473 pKa = 11.15GDD475 pKa = 3.91TDD477 pKa = 3.93TANDD481 pKa = 3.71TYY483 pKa = 10.93FYY485 pKa = 8.94TNEE488 pKa = 3.65YY489 pKa = 9.88GFYY492 pKa = 10.03GTTTPSDD499 pKa = 3.37NTGTDD504 pKa = 3.67TINAAAITSNSYY516 pKa = 11.36LDD518 pKa = 4.58LNAGSTSILNGTTLIINTGTIIEE541 pKa = 4.29NAFGGDD547 pKa = 3.62GNDD550 pKa = 3.74TIIGNSAANVLSGGRR565 pKa = 11.84GDD567 pKa = 5.05DD568 pKa = 3.99SLDD571 pKa = 3.53GGAGNDD577 pKa = 3.64TLRR580 pKa = 11.84GGRR583 pKa = 11.84DD584 pKa = 2.99NDD586 pKa = 3.92TYY588 pKa = 11.13IVNTSGDD595 pKa = 3.85VVIEE599 pKa = 3.99NANEE603 pKa = 4.59GIDD606 pKa = 3.88TIRR609 pKa = 11.84TSISYY614 pKa = 8.57TLGANLEE621 pKa = 4.04NLILTGTAAINGTGNTLNNSITGNSGNNTLNGGVGNDD658 pKa = 3.93TLNGGAGNDD667 pKa = 3.73TYY669 pKa = 11.14TVDD672 pKa = 3.67SIGDD676 pKa = 3.69VVIEE680 pKa = 4.12NANEE684 pKa = 3.94GTDD687 pKa = 3.75TIRR690 pKa = 11.84TSISYY695 pKa = 8.62TLGSNLEE702 pKa = 3.95NLILTGTAAINGTGNTLNNSITGNSGNNTLNGGAGNDD739 pKa = 3.73TLTAGAGNDD748 pKa = 3.65TLTGGLGRR756 pKa = 11.84DD757 pKa = 3.08SFNFNSRR764 pKa = 11.84TEE766 pKa = 4.46GIDD769 pKa = 3.41RR770 pKa = 11.84ITDD773 pKa = 3.8FSVIDD778 pKa = 3.5DD779 pKa = 4.61TIFVSASGFGGGLVTGAAIAANRR802 pKa = 11.84FVIGAAATTSSQRR815 pKa = 11.84FIYY818 pKa = 10.88NNVTGGLFFDD828 pKa = 4.87ADD830 pKa = 3.73GTGAIARR837 pKa = 11.84IQFATLSAGLSLTNADD853 pKa = 3.29IFVNTT858 pKa = 4.48
Molecular weight: 89.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9QFD9|K9QFD9_9NOSO Formyltetrahydrofolate deformylase OS=Nostoc sp. PCC 7107 OX=317936 GN=purU PE=3 SV=1
MM1 pKa = 7.12SQSSLNRR8 pKa = 11.84RR9 pKa = 11.84LTQAFGILLGIGIAIWILRR28 pKa = 11.84GLGILTFLPGGIIWLFVLGAIALGIISFAQRR59 pKa = 11.84TWWRR63 pKa = 11.84FF64 pKa = 2.98
MM1 pKa = 7.12SQSSLNRR8 pKa = 11.84RR9 pKa = 11.84LTQAFGILLGIGIAIWILRR28 pKa = 11.84GLGILTFLPGGIIWLFVLGAIALGIISFAQRR59 pKa = 11.84TWWRR63 pKa = 11.84FF64 pKa = 2.98
Molecular weight: 7.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1692159 |
31 |
2335 |
326.4 |
36.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.236 ± 0.037 | 0.965 ± 0.011 |
4.785 ± 0.026 | 6.096 ± 0.032 |
3.923 ± 0.02 | 6.508 ± 0.032 |
1.828 ± 0.015 | 7.033 ± 0.029 |
4.844 ± 0.03 | 11.017 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.787 ± 0.017 | 4.619 ± 0.033 |
4.629 ± 0.027 | 5.623 ± 0.03 |
4.891 ± 0.027 | 6.322 ± 0.029 |
5.8 ± 0.029 | 6.595 ± 0.028 |
1.392 ± 0.015 | 3.108 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
