
Tetragenococcus muriaticus 3MR10-3
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Enterococcaceae; Tetragenococcus; Tetragenococcus muriaticus
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
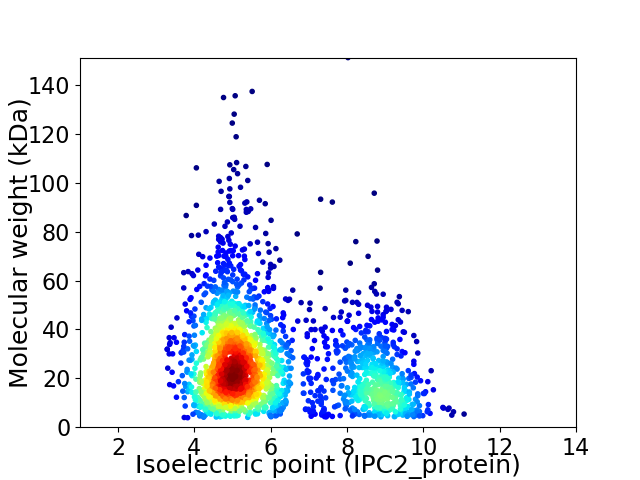
Virtual 2D-PAGE plot for 2262 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A091C807|A0A091C807_9ENTE Di/tripeptide permease OS=Tetragenococcus muriaticus 3MR10-3 OX=1302648 GN=TMU3MR103_0517 PE=3 SV=1
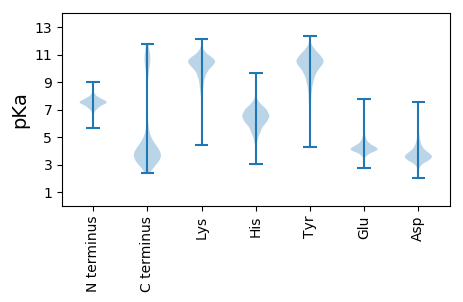
MM1 pKa = 7.59APYY4 pKa = 10.44DD5 pKa = 3.66ATEE8 pKa = 4.05EE9 pKa = 4.19TGSVEE14 pKa = 4.24TQNDD18 pKa = 3.36EE19 pKa = 4.77TVTEE23 pKa = 4.38DD24 pKa = 3.69ANTEE28 pKa = 4.07PEE30 pKa = 4.24VNRR33 pKa = 11.84SYY35 pKa = 11.6SEE37 pKa = 4.17TEE39 pKa = 3.76EE40 pKa = 4.07QQPSSAEE47 pKa = 4.02DD48 pKa = 3.27QTSYY52 pKa = 11.65AQGQEE57 pKa = 3.94QSQTADD63 pKa = 2.96EE64 pKa = 4.47RR65 pKa = 11.84SQLEE69 pKa = 4.04EE70 pKa = 3.75QQEE73 pKa = 4.56GEE75 pKa = 4.13EE76 pKa = 4.45APAEE80 pKa = 4.07EE81 pKa = 4.73DD82 pKa = 2.96EE83 pKa = 4.55QYY85 pKa = 11.76ANEE88 pKa = 4.03EE89 pKa = 3.95DD90 pKa = 3.86QIAEE94 pKa = 4.21EE95 pKa = 4.21EE96 pKa = 4.27AQTTEE101 pKa = 4.19EE102 pKa = 4.45EE103 pKa = 4.12EE104 pKa = 4.53TYY106 pKa = 11.06DD107 pKa = 3.19ATEE110 pKa = 3.82EE111 pKa = 4.09MEE113 pKa = 4.34EE114 pKa = 4.43LEE116 pKa = 4.96LPEE119 pKa = 6.64DD120 pKa = 4.04EE121 pKa = 5.55DD122 pKa = 3.74AQLQEE127 pKa = 4.97EE128 pKa = 4.98VGQHH132 pKa = 6.22PGDD135 pKa = 3.72NNGPAVSTISSNAPEE150 pKa = 3.88PSAFIEE156 pKa = 4.2EE157 pKa = 4.39LAGHH161 pKa = 6.87AEE163 pKa = 4.23SVAAANNLYY172 pKa = 10.26ASVMIAQAVVEE183 pKa = 4.76SGWGPSTLSQAPNNNLFGIKK203 pKa = 8.58GTYY206 pKa = 9.41NGQSVKK212 pKa = 10.14MPTQEE217 pKa = 4.42YY218 pKa = 9.67INGRR222 pKa = 11.84YY223 pKa = 6.78VTVNADD229 pKa = 3.19FRR231 pKa = 11.84KK232 pKa = 9.07YY233 pKa = 10.31PSYY236 pKa = 9.78TASFQDD242 pKa = 3.43NAALLSTNLYY252 pKa = 9.79SGAWKK257 pKa = 10.76SNTSSYY263 pKa = 11.18RR264 pKa = 11.84DD265 pKa = 3.25ATAALTGLYY274 pKa = 8.38ATAPNYY280 pKa = 8.26NTVLNGVIEE289 pKa = 4.97DD290 pKa = 4.0YY291 pKa = 11.32NLTRR295 pKa = 11.84FDD297 pKa = 4.02TGDD300 pKa = 3.14SDD302 pKa = 5.79GIIDD306 pKa = 4.13TGTGNDD312 pKa = 3.89GNSSEE317 pKa = 5.27DD318 pKa = 3.65NNNEE322 pKa = 3.95DD323 pKa = 3.77NSNNEE328 pKa = 3.89NSSSNSGSNTYY339 pKa = 7.27TVRR342 pKa = 11.84PGDD345 pKa = 4.42SVWLIANNNGISMDD359 pKa = 4.5QLRR362 pKa = 11.84SWNNIQNDD370 pKa = 3.82FVYY373 pKa = 10.07PGQEE377 pKa = 3.84LTVSQGNNSSGSNNSNNSGSSNDD400 pKa = 3.75NNNSSSSGSDD410 pKa = 2.9TYY412 pKa = 9.58TVRR415 pKa = 11.84SGDD418 pKa = 4.32SVWLIANNNGISMDD432 pKa = 4.5QLRR435 pKa = 11.84SWNNIQNDD443 pKa = 3.82FVYY446 pKa = 10.07PGQEE450 pKa = 3.84LTVSQGNSSSGSNNSNSGGSSNSGNNSGGSYY481 pKa = 7.52TVRR484 pKa = 11.84SGDD487 pKa = 4.22SVWLIANNNGISMDD501 pKa = 4.5QLRR504 pKa = 11.84SWNNIQNDD512 pKa = 3.89FVYY515 pKa = 10.17PGQTLTVNNGSSSNRR530 pKa = 11.84NTNQSNNSSNNNHH543 pKa = 6.44RR544 pKa = 11.84VTSGDD549 pKa = 4.06SLWMISQEE557 pKa = 3.88YY558 pKa = 9.92GIAISQLKK566 pKa = 10.05SINNLNSDD574 pKa = 4.39TIFIGQTLRR583 pKa = 11.84VSS585 pKa = 3.47
MM1 pKa = 7.59APYY4 pKa = 10.44DD5 pKa = 3.66ATEE8 pKa = 4.05EE9 pKa = 4.19TGSVEE14 pKa = 4.24TQNDD18 pKa = 3.36EE19 pKa = 4.77TVTEE23 pKa = 4.38DD24 pKa = 3.69ANTEE28 pKa = 4.07PEE30 pKa = 4.24VNRR33 pKa = 11.84SYY35 pKa = 11.6SEE37 pKa = 4.17TEE39 pKa = 3.76EE40 pKa = 4.07QQPSSAEE47 pKa = 4.02DD48 pKa = 3.27QTSYY52 pKa = 11.65AQGQEE57 pKa = 3.94QSQTADD63 pKa = 2.96EE64 pKa = 4.47RR65 pKa = 11.84SQLEE69 pKa = 4.04EE70 pKa = 3.75QQEE73 pKa = 4.56GEE75 pKa = 4.13EE76 pKa = 4.45APAEE80 pKa = 4.07EE81 pKa = 4.73DD82 pKa = 2.96EE83 pKa = 4.55QYY85 pKa = 11.76ANEE88 pKa = 4.03EE89 pKa = 3.95DD90 pKa = 3.86QIAEE94 pKa = 4.21EE95 pKa = 4.21EE96 pKa = 4.27AQTTEE101 pKa = 4.19EE102 pKa = 4.45EE103 pKa = 4.12EE104 pKa = 4.53TYY106 pKa = 11.06DD107 pKa = 3.19ATEE110 pKa = 3.82EE111 pKa = 4.09MEE113 pKa = 4.34EE114 pKa = 4.43LEE116 pKa = 4.96LPEE119 pKa = 6.64DD120 pKa = 4.04EE121 pKa = 5.55DD122 pKa = 3.74AQLQEE127 pKa = 4.97EE128 pKa = 4.98VGQHH132 pKa = 6.22PGDD135 pKa = 3.72NNGPAVSTISSNAPEE150 pKa = 3.88PSAFIEE156 pKa = 4.2EE157 pKa = 4.39LAGHH161 pKa = 6.87AEE163 pKa = 4.23SVAAANNLYY172 pKa = 10.26ASVMIAQAVVEE183 pKa = 4.76SGWGPSTLSQAPNNNLFGIKK203 pKa = 8.58GTYY206 pKa = 9.41NGQSVKK212 pKa = 10.14MPTQEE217 pKa = 4.42YY218 pKa = 9.67INGRR222 pKa = 11.84YY223 pKa = 6.78VTVNADD229 pKa = 3.19FRR231 pKa = 11.84KK232 pKa = 9.07YY233 pKa = 10.31PSYY236 pKa = 9.78TASFQDD242 pKa = 3.43NAALLSTNLYY252 pKa = 9.79SGAWKK257 pKa = 10.76SNTSSYY263 pKa = 11.18RR264 pKa = 11.84DD265 pKa = 3.25ATAALTGLYY274 pKa = 8.38ATAPNYY280 pKa = 8.26NTVLNGVIEE289 pKa = 4.97DD290 pKa = 4.0YY291 pKa = 11.32NLTRR295 pKa = 11.84FDD297 pKa = 4.02TGDD300 pKa = 3.14SDD302 pKa = 5.79GIIDD306 pKa = 4.13TGTGNDD312 pKa = 3.89GNSSEE317 pKa = 5.27DD318 pKa = 3.65NNNEE322 pKa = 3.95DD323 pKa = 3.77NSNNEE328 pKa = 3.89NSSSNSGSNTYY339 pKa = 7.27TVRR342 pKa = 11.84PGDD345 pKa = 4.42SVWLIANNNGISMDD359 pKa = 4.5QLRR362 pKa = 11.84SWNNIQNDD370 pKa = 3.82FVYY373 pKa = 10.07PGQEE377 pKa = 3.84LTVSQGNNSSGSNNSNNSGSSNDD400 pKa = 3.75NNNSSSSGSDD410 pKa = 2.9TYY412 pKa = 9.58TVRR415 pKa = 11.84SGDD418 pKa = 4.32SVWLIANNNGISMDD432 pKa = 4.5QLRR435 pKa = 11.84SWNNIQNDD443 pKa = 3.82FVYY446 pKa = 10.07PGQEE450 pKa = 3.84LTVSQGNSSSGSNNSNSGGSSNSGNNSGGSYY481 pKa = 7.52TVRR484 pKa = 11.84SGDD487 pKa = 4.22SVWLIANNNGISMDD501 pKa = 4.5QLRR504 pKa = 11.84SWNNIQNDD512 pKa = 3.89FVYY515 pKa = 10.17PGQTLTVNNGSSSNRR530 pKa = 11.84NTNQSNNSSNNNHH543 pKa = 6.44RR544 pKa = 11.84VTSGDD549 pKa = 4.06SLWMISQEE557 pKa = 3.88YY558 pKa = 9.92GIAISQLKK566 pKa = 10.05SINNLNSDD574 pKa = 4.39TIFIGQTLRR583 pKa = 11.84VSS585 pKa = 3.47
Molecular weight: 63.22 kDa
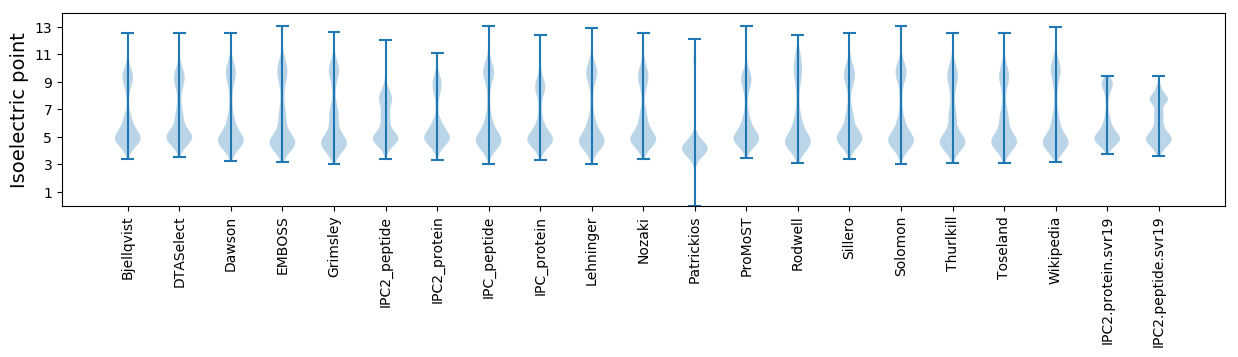
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A091BWM5|A0A091BWM5_9ENTE DNA gyrase subunit B OS=Tetragenococcus muriaticus 3MR10-3 OX=1302648 GN=gyrB PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.24QPNKK9 pKa = 9.14RR10 pKa = 11.84KK11 pKa = 8.54HH12 pKa = 5.23QKK14 pKa = 8.76VHH16 pKa = 5.58GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTKK25 pKa = 10.16NGRR28 pKa = 11.84HH29 pKa = 4.05VLANRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.35GRR39 pKa = 11.84KK40 pKa = 8.82KK41 pKa = 10.58LSAA44 pKa = 3.95
MM1 pKa = 7.35KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 10.24QPNKK9 pKa = 9.14RR10 pKa = 11.84KK11 pKa = 8.54HH12 pKa = 5.23QKK14 pKa = 8.76VHH16 pKa = 5.58GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTKK25 pKa = 10.16NGRR28 pKa = 11.84HH29 pKa = 4.05VLANRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.35GRR39 pKa = 11.84KK40 pKa = 8.82KK41 pKa = 10.58LSAA44 pKa = 3.95
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
565595 |
37 |
1424 |
250.0 |
28.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.871 ± 0.046 | 0.572 ± 0.014 |
5.66 ± 0.046 | 7.806 ± 0.067 |
4.575 ± 0.044 | 6.399 ± 0.054 |
1.836 ± 0.027 | 7.435 ± 0.05 |
6.564 ± 0.054 | 9.647 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.683 ± 0.024 | 4.85 ± 0.039 |
3.371 ± 0.028 | 4.775 ± 0.051 |
3.923 ± 0.038 | 5.974 ± 0.041 |
5.75 ± 0.032 | 6.718 ± 0.041 |
0.914 ± 0.019 | 3.676 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
