
Penicillium sp. occitanis
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Penicillium; unclassified Penicillium
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
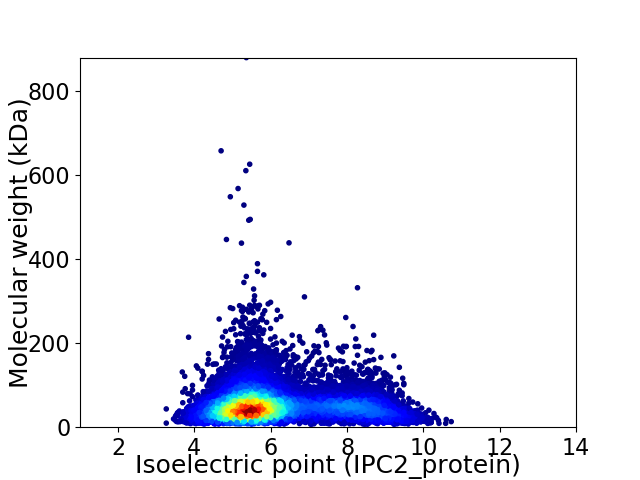
Virtual 2D-PAGE plot for 11231 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H3I571|A0A2H3I571_9EURO Uncharacterized protein OS=Penicillium sp. 'occitanis' OX=290292 GN=PENO1_079590 PE=4 SV=1
MM1 pKa = 7.86PSLTQVSLFLSLTIPGLVQAVPAPAPTTPPKK32 pKa = 10.35LSEE35 pKa = 4.09RR36 pKa = 11.84ASSCTFSGSQGAASASKK53 pKa = 10.73SKK55 pKa = 9.52TSCSTIVLSDD65 pKa = 3.44VAVPSGTTLDD75 pKa = 3.9LTDD78 pKa = 5.36LNDD81 pKa = 3.68GTSVIFEE88 pKa = 4.31GTTSFGYY95 pKa = 9.98EE96 pKa = 3.81EE97 pKa = 4.47WDD99 pKa = 3.72GPLISVSGADD109 pKa = 3.27ITVTGADD116 pKa = 3.33GHH118 pKa = 6.8VIDD121 pKa = 5.78GDD123 pKa = 4.0GSRR126 pKa = 11.84WWDD129 pKa = 3.42GEE131 pKa = 3.95GSNGGTTKK139 pKa = 10.62PKK141 pKa = 10.28FFYY144 pKa = 10.74AHH146 pKa = 7.7DD147 pKa = 4.24LTSSTISGLSLKK159 pKa = 10.27NSPVQTFSIDD169 pKa = 2.93GSTDD173 pKa = 3.11LTLSDD178 pKa = 3.63ITIDD182 pKa = 5.56DD183 pKa = 3.68SDD185 pKa = 5.41GDD187 pKa = 4.34DD188 pKa = 3.62GSAANTDD195 pKa = 3.33AFDD198 pKa = 3.59VGEE201 pKa = 4.22STGIIISGATVYY213 pKa = 10.98NQDD216 pKa = 3.5DD217 pKa = 4.0CLAINSGTNITFTGGYY233 pKa = 9.4CSGGHH238 pKa = 5.92GLSIGSVGGRR248 pKa = 11.84DD249 pKa = 3.83DD250 pKa = 3.76NTVSGVTIEE259 pKa = 4.66SSTITNSANGVRR271 pKa = 11.84IKK273 pKa = 9.75TVYY276 pKa = 10.4DD277 pKa = 3.22ATGSVTGVTYY287 pKa = 11.04KK288 pKa = 10.86DD289 pKa = 3.11ITLSGITDD297 pKa = 3.45YY298 pKa = 11.69GIVIEE303 pKa = 4.22QDD305 pKa = 3.68YY306 pKa = 11.2EE307 pKa = 4.24NGSPTGTPTTGVPITDD323 pKa = 3.59LTIDD327 pKa = 3.64NVQGTVEE334 pKa = 3.96SDD336 pKa = 2.77ATEE339 pKa = 4.04IYY341 pKa = 9.99ILCGDD346 pKa = 4.84GSCSDD351 pKa = 3.67WTWTDD356 pKa = 3.16VSVTGGEE363 pKa = 4.19TSDD366 pKa = 3.54KK367 pKa = 11.14CEE369 pKa = 4.17NAPDD373 pKa = 5.6DD374 pKa = 4.02ISCC377 pKa = 4.64
MM1 pKa = 7.86PSLTQVSLFLSLTIPGLVQAVPAPAPTTPPKK32 pKa = 10.35LSEE35 pKa = 4.09RR36 pKa = 11.84ASSCTFSGSQGAASASKK53 pKa = 10.73SKK55 pKa = 9.52TSCSTIVLSDD65 pKa = 3.44VAVPSGTTLDD75 pKa = 3.9LTDD78 pKa = 5.36LNDD81 pKa = 3.68GTSVIFEE88 pKa = 4.31GTTSFGYY95 pKa = 9.98EE96 pKa = 3.81EE97 pKa = 4.47WDD99 pKa = 3.72GPLISVSGADD109 pKa = 3.27ITVTGADD116 pKa = 3.33GHH118 pKa = 6.8VIDD121 pKa = 5.78GDD123 pKa = 4.0GSRR126 pKa = 11.84WWDD129 pKa = 3.42GEE131 pKa = 3.95GSNGGTTKK139 pKa = 10.62PKK141 pKa = 10.28FFYY144 pKa = 10.74AHH146 pKa = 7.7DD147 pKa = 4.24LTSSTISGLSLKK159 pKa = 10.27NSPVQTFSIDD169 pKa = 2.93GSTDD173 pKa = 3.11LTLSDD178 pKa = 3.63ITIDD182 pKa = 5.56DD183 pKa = 3.68SDD185 pKa = 5.41GDD187 pKa = 4.34DD188 pKa = 3.62GSAANTDD195 pKa = 3.33AFDD198 pKa = 3.59VGEE201 pKa = 4.22STGIIISGATVYY213 pKa = 10.98NQDD216 pKa = 3.5DD217 pKa = 4.0CLAINSGTNITFTGGYY233 pKa = 9.4CSGGHH238 pKa = 5.92GLSIGSVGGRR248 pKa = 11.84DD249 pKa = 3.83DD250 pKa = 3.76NTVSGVTIEE259 pKa = 4.66SSTITNSANGVRR271 pKa = 11.84IKK273 pKa = 9.75TVYY276 pKa = 10.4DD277 pKa = 3.22ATGSVTGVTYY287 pKa = 11.04KK288 pKa = 10.86DD289 pKa = 3.11ITLSGITDD297 pKa = 3.45YY298 pKa = 11.69GIVIEE303 pKa = 4.22QDD305 pKa = 3.68YY306 pKa = 11.2EE307 pKa = 4.24NGSPTGTPTTGVPITDD323 pKa = 3.59LTIDD327 pKa = 3.64NVQGTVEE334 pKa = 3.96SDD336 pKa = 2.77ATEE339 pKa = 4.04IYY341 pKa = 9.99ILCGDD346 pKa = 4.84GSCSDD351 pKa = 3.67WTWTDD356 pKa = 3.16VSVTGGEE363 pKa = 4.19TSDD366 pKa = 3.54KK367 pKa = 11.14CEE369 pKa = 4.17NAPDD373 pKa = 5.6DD374 pKa = 4.02ISCC377 pKa = 4.64
Molecular weight: 38.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H3I952|A0A2H3I952_9EURO Mitochondrial export protein Som1 OS=Penicillium sp. 'occitanis' OX=290292 GN=PENO1_064200 PE=4 SV=1
MM1 pKa = 7.7LLRR4 pKa = 11.84YY5 pKa = 8.21TEE7 pKa = 4.3LGSRR11 pKa = 11.84RR12 pKa = 11.84TKK14 pKa = 9.22MMEE17 pKa = 3.85YY18 pKa = 10.45VGTQLFWSQSEE29 pKa = 3.84ARR31 pKa = 11.84QAGDD35 pKa = 3.17HH36 pKa = 6.84RR37 pKa = 11.84DD38 pKa = 3.21AKK40 pKa = 9.61TAAEE44 pKa = 3.73ARR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84KK50 pKa = 8.79VQNRR54 pKa = 11.84KK55 pKa = 6.05NQRR58 pKa = 11.84VRR60 pKa = 11.84RR61 pKa = 11.84LRR63 pKa = 11.84LKK65 pKa = 11.0GGDD68 pKa = 2.87AGNFQEE74 pKa = 4.82SRR76 pKa = 11.84PFRR79 pKa = 11.84VKK81 pKa = 9.87RR82 pKa = 11.84WRR84 pKa = 11.84LDD86 pKa = 3.59EE87 pKa = 5.08PDD89 pKa = 5.31HH90 pKa = 6.74IPSSEE95 pKa = 3.89NSPASEE101 pKa = 3.99RR102 pKa = 11.84ATTTAFFHH110 pKa = 6.54GPPHH114 pKa = 6.84AGITASAAKK123 pKa = 9.93GRR125 pKa = 11.84VVLRR129 pKa = 11.84EE130 pKa = 3.9SLSTITHH137 pKa = 5.56SQPLVNLDD145 pKa = 3.27IQPFTFPFSSDD156 pKa = 2.43HH157 pKa = 6.44HH158 pKa = 6.56LLHH161 pKa = 7.07FIHH164 pKa = 6.7YY165 pKa = 7.53NVCRR169 pKa = 11.84ALITNMRR176 pKa = 11.84TLNTVPADD184 pKa = 3.54STFCTIASPCRR195 pKa = 11.84DD196 pKa = 3.8DD197 pKa = 3.52TTLFPLKK204 pKa = 10.21PDD206 pKa = 3.93IPPSLIPTAFQQTHH220 pKa = 5.32DD221 pKa = 3.33HH222 pKa = 6.53FARR225 pKa = 11.84IKK227 pKa = 10.45SFPSLASARR236 pKa = 11.84II237 pKa = 3.59
MM1 pKa = 7.7LLRR4 pKa = 11.84YY5 pKa = 8.21TEE7 pKa = 4.3LGSRR11 pKa = 11.84RR12 pKa = 11.84TKK14 pKa = 9.22MMEE17 pKa = 3.85YY18 pKa = 10.45VGTQLFWSQSEE29 pKa = 3.84ARR31 pKa = 11.84QAGDD35 pKa = 3.17HH36 pKa = 6.84RR37 pKa = 11.84DD38 pKa = 3.21AKK40 pKa = 9.61TAAEE44 pKa = 3.73ARR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84KK50 pKa = 8.79VQNRR54 pKa = 11.84KK55 pKa = 6.05NQRR58 pKa = 11.84VRR60 pKa = 11.84RR61 pKa = 11.84LRR63 pKa = 11.84LKK65 pKa = 11.0GGDD68 pKa = 2.87AGNFQEE74 pKa = 4.82SRR76 pKa = 11.84PFRR79 pKa = 11.84VKK81 pKa = 9.87RR82 pKa = 11.84WRR84 pKa = 11.84LDD86 pKa = 3.59EE87 pKa = 5.08PDD89 pKa = 5.31HH90 pKa = 6.74IPSSEE95 pKa = 3.89NSPASEE101 pKa = 3.99RR102 pKa = 11.84ATTTAFFHH110 pKa = 6.54GPPHH114 pKa = 6.84AGITASAAKK123 pKa = 9.93GRR125 pKa = 11.84VVLRR129 pKa = 11.84EE130 pKa = 3.9SLSTITHH137 pKa = 5.56SQPLVNLDD145 pKa = 3.27IQPFTFPFSSDD156 pKa = 2.43HH157 pKa = 6.44HH158 pKa = 6.56LLHH161 pKa = 7.07FIHH164 pKa = 6.7YY165 pKa = 7.53NVCRR169 pKa = 11.84ALITNMRR176 pKa = 11.84TLNTVPADD184 pKa = 3.54STFCTIASPCRR195 pKa = 11.84DD196 pKa = 3.8DD197 pKa = 3.52TTLFPLKK204 pKa = 10.21PDD206 pKa = 3.93IPPSLIPTAFQQTHH220 pKa = 5.32DD221 pKa = 3.33HH222 pKa = 6.53FARR225 pKa = 11.84IKK227 pKa = 10.45SFPSLASARR236 pKa = 11.84II237 pKa = 3.59
Molecular weight: 26.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5743013 |
66 |
7905 |
511.4 |
56.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.343 ± 0.019 | 1.125 ± 0.007 |
5.732 ± 0.014 | 6.189 ± 0.022 |
3.856 ± 0.012 | 6.674 ± 0.02 |
2.345 ± 0.009 | 5.446 ± 0.014 |
4.768 ± 0.019 | 9.039 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.144 ± 0.009 | 3.966 ± 0.011 |
5.587 ± 0.022 | 4.012 ± 0.015 |
5.711 ± 0.019 | 8.214 ± 0.022 |
6.162 ± 0.014 | 6.223 ± 0.014 |
1.485 ± 0.009 | 2.979 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
