
Festuca pratensis amalgavirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; unclassified Amalgaviridae
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
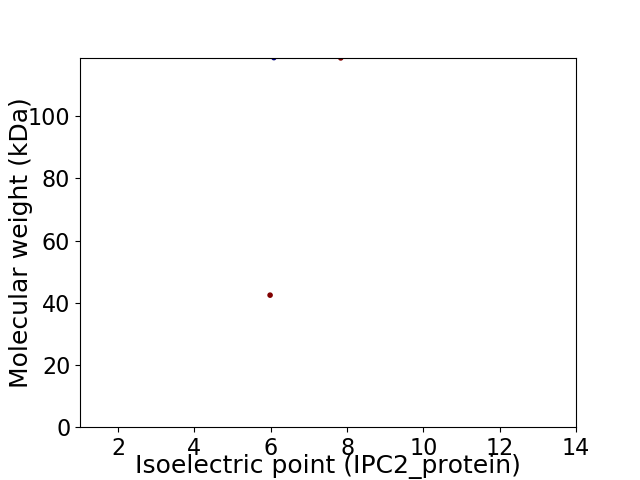
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2Z6JIN1|A0A2Z6JIN1_9VIRU ORF1p OS=Festuca pratensis amalgavirus 2 OX=2069325 GN=ORF1 PE=4 SV=1
MM1 pKa = 7.35SVDD4 pKa = 3.53PLRR7 pKa = 11.84FLEE10 pKa = 4.96DD11 pKa = 2.99AATAQAADD19 pKa = 4.45AIKK22 pKa = 10.42LRR24 pKa = 11.84PLMLEE29 pKa = 4.08LSGSKK34 pKa = 10.2VLAKK38 pKa = 10.48EE39 pKa = 3.82CDD41 pKa = 3.26IEE43 pKa = 4.27HH44 pKa = 7.07LLNAGFTVDD53 pKa = 5.38DD54 pKa = 4.04VEE56 pKa = 5.02KK57 pKa = 10.58LSKK60 pKa = 10.3SLKK63 pKa = 9.46PLIDD67 pKa = 3.1QGVFDD72 pKa = 5.69DD73 pKa = 4.28AWTMAKK79 pKa = 10.2GSGIILSAQEE89 pKa = 3.87MTFPDD94 pKa = 3.71LFMFRR99 pKa = 11.84RR100 pKa = 11.84WLTTPQGAQALSLVQARR117 pKa = 11.84RR118 pKa = 11.84KK119 pKa = 5.79MTKK122 pKa = 9.78AGKK125 pKa = 9.67KK126 pKa = 9.5VLGHH130 pKa = 6.0QDD132 pKa = 3.07VALLRR137 pKa = 11.84LLQHH141 pKa = 6.36YY142 pKa = 9.86EE143 pKa = 3.52DD144 pKa = 4.15DD145 pKa = 4.23ARR147 pKa = 11.84RR148 pKa = 11.84EE149 pKa = 3.75LDD151 pKa = 3.14AKK153 pKa = 10.49RR154 pKa = 11.84VEE156 pKa = 4.3TEE158 pKa = 3.62AAVAQLQAEE167 pKa = 4.09IDD169 pKa = 3.8KK170 pKa = 11.04LKK172 pKa = 10.82KK173 pKa = 10.28KK174 pKa = 9.34YY175 pKa = 10.74AKK177 pKa = 10.31AEE179 pKa = 4.13KK180 pKa = 9.31KK181 pKa = 9.8QKK183 pKa = 10.41RR184 pKa = 11.84DD185 pKa = 3.37FPLIANYY192 pKa = 9.96VPLTDD197 pKa = 3.91NEE199 pKa = 4.19VRR201 pKa = 11.84NQAWDD206 pKa = 3.9MYY208 pKa = 9.91CQQCINEE215 pKa = 4.36GSVPMGRR222 pKa = 11.84TSTNLKK228 pKa = 9.96IVGDD232 pKa = 3.93KK233 pKa = 10.78FRR235 pKa = 11.84DD236 pKa = 4.02HH237 pKa = 7.06IVQAHH242 pKa = 6.75KK243 pKa = 9.63LTYY246 pKa = 9.67CQQPDD251 pKa = 3.42HH252 pKa = 6.87TDD254 pKa = 2.92ALIEE258 pKa = 4.24FGKK261 pKa = 10.49QKK263 pKa = 10.34ILRR266 pKa = 11.84CQEE269 pKa = 4.0AGSGKK274 pKa = 10.31LEE276 pKa = 3.95SSFRR280 pKa = 11.84NLLTVLNPEE289 pKa = 4.22NAATLPPRR297 pKa = 11.84TEE299 pKa = 3.92EE300 pKa = 4.38ADD302 pKa = 3.81DD303 pKa = 5.18GVDD306 pKa = 3.44TDD308 pKa = 4.68GGSPNPPEE316 pKa = 5.17PDD318 pKa = 2.95THH320 pKa = 8.17ADD322 pKa = 3.53AEE324 pKa = 4.36PAADD328 pKa = 3.61QPTPPEE334 pKa = 3.8ATGPGSSAGPTPPQSSRR351 pKa = 11.84SHH353 pKa = 5.98KK354 pKa = 9.89RR355 pKa = 11.84KK356 pKa = 9.63QPAVDD361 pKa = 3.79AQSEE365 pKa = 4.38DD366 pKa = 3.76PNPSKK371 pKa = 10.72KK372 pKa = 10.08RR373 pKa = 11.84STRR376 pKa = 11.84AGRR379 pKa = 11.84HH380 pKa = 4.91PNRR383 pKa = 11.84AEE385 pKa = 3.69
MM1 pKa = 7.35SVDD4 pKa = 3.53PLRR7 pKa = 11.84FLEE10 pKa = 4.96DD11 pKa = 2.99AATAQAADD19 pKa = 4.45AIKK22 pKa = 10.42LRR24 pKa = 11.84PLMLEE29 pKa = 4.08LSGSKK34 pKa = 10.2VLAKK38 pKa = 10.48EE39 pKa = 3.82CDD41 pKa = 3.26IEE43 pKa = 4.27HH44 pKa = 7.07LLNAGFTVDD53 pKa = 5.38DD54 pKa = 4.04VEE56 pKa = 5.02KK57 pKa = 10.58LSKK60 pKa = 10.3SLKK63 pKa = 9.46PLIDD67 pKa = 3.1QGVFDD72 pKa = 5.69DD73 pKa = 4.28AWTMAKK79 pKa = 10.2GSGIILSAQEE89 pKa = 3.87MTFPDD94 pKa = 3.71LFMFRR99 pKa = 11.84RR100 pKa = 11.84WLTTPQGAQALSLVQARR117 pKa = 11.84RR118 pKa = 11.84KK119 pKa = 5.79MTKK122 pKa = 9.78AGKK125 pKa = 9.67KK126 pKa = 9.5VLGHH130 pKa = 6.0QDD132 pKa = 3.07VALLRR137 pKa = 11.84LLQHH141 pKa = 6.36YY142 pKa = 9.86EE143 pKa = 3.52DD144 pKa = 4.15DD145 pKa = 4.23ARR147 pKa = 11.84RR148 pKa = 11.84EE149 pKa = 3.75LDD151 pKa = 3.14AKK153 pKa = 10.49RR154 pKa = 11.84VEE156 pKa = 4.3TEE158 pKa = 3.62AAVAQLQAEE167 pKa = 4.09IDD169 pKa = 3.8KK170 pKa = 11.04LKK172 pKa = 10.82KK173 pKa = 10.28KK174 pKa = 9.34YY175 pKa = 10.74AKK177 pKa = 10.31AEE179 pKa = 4.13KK180 pKa = 9.31KK181 pKa = 9.8QKK183 pKa = 10.41RR184 pKa = 11.84DD185 pKa = 3.37FPLIANYY192 pKa = 9.96VPLTDD197 pKa = 3.91NEE199 pKa = 4.19VRR201 pKa = 11.84NQAWDD206 pKa = 3.9MYY208 pKa = 9.91CQQCINEE215 pKa = 4.36GSVPMGRR222 pKa = 11.84TSTNLKK228 pKa = 9.96IVGDD232 pKa = 3.93KK233 pKa = 10.78FRR235 pKa = 11.84DD236 pKa = 4.02HH237 pKa = 7.06IVQAHH242 pKa = 6.75KK243 pKa = 9.63LTYY246 pKa = 9.67CQQPDD251 pKa = 3.42HH252 pKa = 6.87TDD254 pKa = 2.92ALIEE258 pKa = 4.24FGKK261 pKa = 10.49QKK263 pKa = 10.34ILRR266 pKa = 11.84CQEE269 pKa = 4.0AGSGKK274 pKa = 10.31LEE276 pKa = 3.95SSFRR280 pKa = 11.84NLLTVLNPEE289 pKa = 4.22NAATLPPRR297 pKa = 11.84TEE299 pKa = 3.92EE300 pKa = 4.38ADD302 pKa = 3.81DD303 pKa = 5.18GVDD306 pKa = 3.44TDD308 pKa = 4.68GGSPNPPEE316 pKa = 5.17PDD318 pKa = 2.95THH320 pKa = 8.17ADD322 pKa = 3.53AEE324 pKa = 4.36PAADD328 pKa = 3.61QPTPPEE334 pKa = 3.8ATGPGSSAGPTPPQSSRR351 pKa = 11.84SHH353 pKa = 5.98KK354 pKa = 9.89RR355 pKa = 11.84KK356 pKa = 9.63QPAVDD361 pKa = 3.79AQSEE365 pKa = 4.38DD366 pKa = 3.76PNPSKK371 pKa = 10.72KK372 pKa = 10.08RR373 pKa = 11.84STRR376 pKa = 11.84AGRR379 pKa = 11.84HH380 pKa = 4.91PNRR383 pKa = 11.84AEE385 pKa = 3.69
Molecular weight: 42.47 kDa
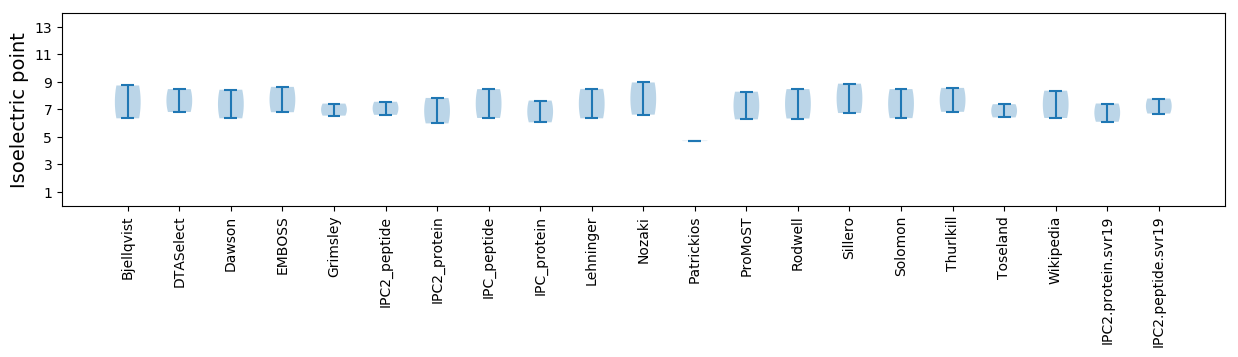
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z6JIN1|A0A2Z6JIN1_9VIRU ORF1p OS=Festuca pratensis amalgavirus 2 OX=2069325 GN=ORF1 PE=4 SV=1
MM1 pKa = 7.35SVDD4 pKa = 3.53PLRR7 pKa = 11.84FLEE10 pKa = 4.96DD11 pKa = 2.99AATAQAADD19 pKa = 4.45AIKK22 pKa = 10.42LRR24 pKa = 11.84PLMLEE29 pKa = 4.08LSGSKK34 pKa = 10.2VLAKK38 pKa = 10.48EE39 pKa = 3.82CDD41 pKa = 3.26IEE43 pKa = 4.27HH44 pKa = 7.07LLNAGFTVDD53 pKa = 5.38DD54 pKa = 4.04VEE56 pKa = 5.02KK57 pKa = 10.58LSKK60 pKa = 10.3SLKK63 pKa = 9.46PLIDD67 pKa = 3.1QGVFDD72 pKa = 5.69DD73 pKa = 4.28AWTMAKK79 pKa = 10.2GSGIILSAQEE89 pKa = 3.87MTFPDD94 pKa = 3.71LFMFRR99 pKa = 11.84RR100 pKa = 11.84WLTTPQGAQALSLVQARR117 pKa = 11.84RR118 pKa = 11.84KK119 pKa = 5.79MTKK122 pKa = 9.78AGKK125 pKa = 9.67KK126 pKa = 9.5VLGHH130 pKa = 6.0QDD132 pKa = 3.07VALLRR137 pKa = 11.84LLQHH141 pKa = 6.36YY142 pKa = 9.86EE143 pKa = 3.52DD144 pKa = 4.15DD145 pKa = 4.23ARR147 pKa = 11.84RR148 pKa = 11.84EE149 pKa = 3.75LDD151 pKa = 3.14AKK153 pKa = 10.49RR154 pKa = 11.84VEE156 pKa = 4.3TEE158 pKa = 3.62AAVAQLQAEE167 pKa = 4.09IDD169 pKa = 3.8KK170 pKa = 11.04LKK172 pKa = 10.82KK173 pKa = 10.28KK174 pKa = 9.34YY175 pKa = 10.74AKK177 pKa = 10.31AEE179 pKa = 4.13KK180 pKa = 9.31KK181 pKa = 9.8QKK183 pKa = 10.41RR184 pKa = 11.84DD185 pKa = 3.37FPLIANYY192 pKa = 9.96VPLTDD197 pKa = 3.91NEE199 pKa = 4.19VRR201 pKa = 11.84NQAWDD206 pKa = 3.9MYY208 pKa = 9.91CQQCINEE215 pKa = 4.36GSVPMGRR222 pKa = 11.84TSTNLKK228 pKa = 9.96IVGDD232 pKa = 3.93KK233 pKa = 10.78FRR235 pKa = 11.84DD236 pKa = 4.02HH237 pKa = 7.06IVQAHH242 pKa = 6.75KK243 pKa = 9.63LTYY246 pKa = 9.67CQQPDD251 pKa = 3.42HH252 pKa = 6.87TDD254 pKa = 2.92ALIEE258 pKa = 4.24FGKK261 pKa = 10.49QKK263 pKa = 10.34ILRR266 pKa = 11.84CQEE269 pKa = 4.0AGSGKK274 pKa = 10.27LEE276 pKa = 4.28SSFVTYY282 pKa = 10.46SLSSIQRR289 pKa = 11.84MLLRR293 pKa = 11.84YY294 pKa = 9.0PLAQRR299 pKa = 11.84KK300 pKa = 7.47QMMEE304 pKa = 5.05WIPMGVVPTRR314 pKa = 11.84PSLTRR319 pKa = 11.84TQMLSQLPTNPLLLKK334 pKa = 10.39PRR336 pKa = 11.84VLGARR341 pKa = 11.84PGPPLPKK348 pKa = 9.97AVEE351 pKa = 4.67AISGNSLLSTPNRR364 pKa = 11.84RR365 pKa = 11.84IQIHH369 pKa = 6.03RR370 pKa = 11.84RR371 pKa = 11.84SGQPGQAGIPTARR384 pKa = 11.84SRR386 pKa = 11.84WEE388 pKa = 3.56AGVRR392 pKa = 11.84HH393 pKa = 6.67IIGGGEE399 pKa = 3.93LLEE402 pKa = 4.09WRR404 pKa = 11.84VDD406 pKa = 3.37NNKK409 pKa = 9.56YY410 pKa = 10.29RR411 pKa = 11.84GGGNLHH417 pKa = 7.22DD418 pKa = 5.07ALLLIGSADD427 pKa = 3.78DD428 pKa = 3.67VTPYY432 pKa = 9.98TALSSLLSVEE442 pKa = 3.9EE443 pKa = 3.8ARR445 pKa = 11.84RR446 pKa = 11.84VLLLPDD452 pKa = 4.17GLAVPDD458 pKa = 4.76GKK460 pKa = 10.69ACCVMKK466 pKa = 10.53QFNDD470 pKa = 3.67DD471 pKa = 3.34ATAGPLLRR479 pKa = 11.84AFGVKK484 pKa = 9.83GKK486 pKa = 10.4YY487 pKa = 8.78GLKK490 pKa = 10.22AAIEE494 pKa = 4.04QFVWGLYY501 pKa = 10.32DD502 pKa = 3.56RR503 pKa = 11.84VGSEE507 pKa = 4.19DD508 pKa = 3.63LKK510 pKa = 11.0PRR512 pKa = 11.84QLPGLLARR520 pKa = 11.84VGYY523 pKa = 7.67RR524 pKa = 11.84TKK526 pKa = 10.96LLDD529 pKa = 3.36MEE531 pKa = 4.67KK532 pKa = 10.61ALKK535 pKa = 9.94KK536 pKa = 9.72IQAVEE541 pKa = 3.78PLGRR545 pKa = 11.84AVMMLDD551 pKa = 3.04ATEE554 pKa = 4.19QCFSSPLFNAISEE567 pKa = 4.55AVTEE571 pKa = 4.06LHH573 pKa = 6.54SNPRR577 pKa = 11.84SGWRR581 pKa = 11.84NYY583 pKa = 9.62LVRR586 pKa = 11.84ASSAWADD593 pKa = 3.14LWDD596 pKa = 3.9EE597 pKa = 4.57LKK599 pKa = 10.87SCGTIVEE606 pKa = 4.5LDD608 pKa = 3.02WSKK611 pKa = 11.06FDD613 pKa = 4.01RR614 pKa = 11.84DD615 pKa = 3.77RR616 pKa = 11.84PAEE619 pKa = 4.6DD620 pKa = 2.75IQFFVDD626 pKa = 4.62VIISCFRR633 pKa = 11.84PKK635 pKa = 10.44NSRR638 pKa = 11.84EE639 pKa = 3.63RR640 pKa = 11.84RR641 pKa = 11.84LLAGYY646 pKa = 10.27RR647 pKa = 11.84KK648 pKa = 9.2MMEE651 pKa = 3.77NALVHH656 pKa = 5.72RR657 pKa = 11.84VMMLDD662 pKa = 3.75DD663 pKa = 4.64GSFFTLEE670 pKa = 3.86GMVPSGSLWTGICDD684 pKa = 3.35TALNIMYY691 pKa = 8.2ITSALRR697 pKa = 11.84KK698 pKa = 9.97LGFDD702 pKa = 4.65DD703 pKa = 4.07STFSPKK709 pKa = 10.26CAGDD713 pKa = 4.26DD714 pKa = 4.03NLTCFQRR721 pKa = 11.84RR722 pKa = 11.84QGLDD726 pKa = 2.88VMEE729 pKa = 4.78RR730 pKa = 11.84LRR732 pKa = 11.84TTLNEE737 pKa = 3.85MFRR740 pKa = 11.84ANIDD744 pKa = 3.3KK745 pKa = 10.77KK746 pKa = 10.94DD747 pKa = 4.53FIVHH751 pKa = 5.56YY752 pKa = 9.38PPYY755 pKa = 10.94AVTTVQACFEE765 pKa = 4.93PGTDD769 pKa = 3.55LSKK772 pKa = 10.44GTSRR776 pKa = 11.84MMDD779 pKa = 3.04KK780 pKa = 11.24AEE782 pKa = 3.99WVPFTGPCPIDD793 pKa = 3.34QAAGRR798 pKa = 11.84SHH800 pKa = 6.25RR801 pKa = 11.84WKK803 pKa = 10.83YY804 pKa = 11.29VFTNKK809 pKa = 9.86PKK811 pKa = 10.24FLANFFLPDD820 pKa = 3.91GKK822 pKa = 9.73PIRR825 pKa = 11.84PAHH828 pKa = 6.75DD829 pKa = 3.76NLEE832 pKa = 4.15KK833 pKa = 10.93LLWPEE838 pKa = 5.16GIHH841 pKa = 6.89EE842 pKa = 4.64SIEE845 pKa = 4.25DD846 pKa = 3.39YY847 pKa = 10.22MAAVLSMVVDD857 pKa = 3.74NPFNHH862 pKa = 6.68HH863 pKa = 5.94NVNHH867 pKa = 5.5MMHH870 pKa = 7.36RR871 pKa = 11.84YY872 pKa = 9.94LIAQQIQKK880 pKa = 10.24QSIFIDD886 pKa = 3.58PAVVMEE892 pKa = 4.45LAKK895 pKa = 10.01TRR897 pKa = 11.84PKK899 pKa = 10.47HH900 pKa = 5.75GEE902 pKa = 3.61AVPFPEE908 pKa = 3.16IAYY911 pKa = 9.24YY912 pKa = 9.86RR913 pKa = 11.84KK914 pKa = 10.02CEE916 pKa = 4.11GYY918 pKa = 10.72VDD920 pKa = 3.9IEE922 pKa = 4.31RR923 pKa = 11.84EE924 pKa = 3.99PEE926 pKa = 3.67FQEE929 pKa = 4.12FFKK932 pKa = 11.02SFRR935 pKa = 11.84EE936 pKa = 4.57FISSVSTLYY945 pKa = 10.78ARR947 pKa = 11.84RR948 pKa = 11.84SEE950 pKa = 4.13GGIDD954 pKa = 2.83AWRR957 pKa = 11.84FMDD960 pKa = 5.13IIRR963 pKa = 11.84GQHH966 pKa = 6.21SIGAGQFGNSVHH978 pKa = 5.32EE979 pKa = 4.07WCRR982 pKa = 11.84FLGEE986 pKa = 4.36HH987 pKa = 6.9PLTRR991 pKa = 11.84SLRR994 pKa = 11.84KK995 pKa = 9.44ARR997 pKa = 11.84RR998 pKa = 11.84FRR1000 pKa = 11.84PMKK1003 pKa = 9.28EE1004 pKa = 3.64AVVADD1009 pKa = 4.24PEE1011 pKa = 4.49TLGKK1015 pKa = 9.95VEE1017 pKa = 5.52KK1018 pKa = 10.85AFTWVLDD1025 pKa = 3.63LCEE1028 pKa = 5.48ANDD1031 pKa = 3.56PMTPLFYY1038 pKa = 10.85ASAVSDD1044 pKa = 3.36ILSSNVSAPP1053 pKa = 3.23
MM1 pKa = 7.35SVDD4 pKa = 3.53PLRR7 pKa = 11.84FLEE10 pKa = 4.96DD11 pKa = 2.99AATAQAADD19 pKa = 4.45AIKK22 pKa = 10.42LRR24 pKa = 11.84PLMLEE29 pKa = 4.08LSGSKK34 pKa = 10.2VLAKK38 pKa = 10.48EE39 pKa = 3.82CDD41 pKa = 3.26IEE43 pKa = 4.27HH44 pKa = 7.07LLNAGFTVDD53 pKa = 5.38DD54 pKa = 4.04VEE56 pKa = 5.02KK57 pKa = 10.58LSKK60 pKa = 10.3SLKK63 pKa = 9.46PLIDD67 pKa = 3.1QGVFDD72 pKa = 5.69DD73 pKa = 4.28AWTMAKK79 pKa = 10.2GSGIILSAQEE89 pKa = 3.87MTFPDD94 pKa = 3.71LFMFRR99 pKa = 11.84RR100 pKa = 11.84WLTTPQGAQALSLVQARR117 pKa = 11.84RR118 pKa = 11.84KK119 pKa = 5.79MTKK122 pKa = 9.78AGKK125 pKa = 9.67KK126 pKa = 9.5VLGHH130 pKa = 6.0QDD132 pKa = 3.07VALLRR137 pKa = 11.84LLQHH141 pKa = 6.36YY142 pKa = 9.86EE143 pKa = 3.52DD144 pKa = 4.15DD145 pKa = 4.23ARR147 pKa = 11.84RR148 pKa = 11.84EE149 pKa = 3.75LDD151 pKa = 3.14AKK153 pKa = 10.49RR154 pKa = 11.84VEE156 pKa = 4.3TEE158 pKa = 3.62AAVAQLQAEE167 pKa = 4.09IDD169 pKa = 3.8KK170 pKa = 11.04LKK172 pKa = 10.82KK173 pKa = 10.28KK174 pKa = 9.34YY175 pKa = 10.74AKK177 pKa = 10.31AEE179 pKa = 4.13KK180 pKa = 9.31KK181 pKa = 9.8QKK183 pKa = 10.41RR184 pKa = 11.84DD185 pKa = 3.37FPLIANYY192 pKa = 9.96VPLTDD197 pKa = 3.91NEE199 pKa = 4.19VRR201 pKa = 11.84NQAWDD206 pKa = 3.9MYY208 pKa = 9.91CQQCINEE215 pKa = 4.36GSVPMGRR222 pKa = 11.84TSTNLKK228 pKa = 9.96IVGDD232 pKa = 3.93KK233 pKa = 10.78FRR235 pKa = 11.84DD236 pKa = 4.02HH237 pKa = 7.06IVQAHH242 pKa = 6.75KK243 pKa = 9.63LTYY246 pKa = 9.67CQQPDD251 pKa = 3.42HH252 pKa = 6.87TDD254 pKa = 2.92ALIEE258 pKa = 4.24FGKK261 pKa = 10.49QKK263 pKa = 10.34ILRR266 pKa = 11.84CQEE269 pKa = 4.0AGSGKK274 pKa = 10.27LEE276 pKa = 4.28SSFVTYY282 pKa = 10.46SLSSIQRR289 pKa = 11.84MLLRR293 pKa = 11.84YY294 pKa = 9.0PLAQRR299 pKa = 11.84KK300 pKa = 7.47QMMEE304 pKa = 5.05WIPMGVVPTRR314 pKa = 11.84PSLTRR319 pKa = 11.84TQMLSQLPTNPLLLKK334 pKa = 10.39PRR336 pKa = 11.84VLGARR341 pKa = 11.84PGPPLPKK348 pKa = 9.97AVEE351 pKa = 4.67AISGNSLLSTPNRR364 pKa = 11.84RR365 pKa = 11.84IQIHH369 pKa = 6.03RR370 pKa = 11.84RR371 pKa = 11.84SGQPGQAGIPTARR384 pKa = 11.84SRR386 pKa = 11.84WEE388 pKa = 3.56AGVRR392 pKa = 11.84HH393 pKa = 6.67IIGGGEE399 pKa = 3.93LLEE402 pKa = 4.09WRR404 pKa = 11.84VDD406 pKa = 3.37NNKK409 pKa = 9.56YY410 pKa = 10.29RR411 pKa = 11.84GGGNLHH417 pKa = 7.22DD418 pKa = 5.07ALLLIGSADD427 pKa = 3.78DD428 pKa = 3.67VTPYY432 pKa = 9.98TALSSLLSVEE442 pKa = 3.9EE443 pKa = 3.8ARR445 pKa = 11.84RR446 pKa = 11.84VLLLPDD452 pKa = 4.17GLAVPDD458 pKa = 4.76GKK460 pKa = 10.69ACCVMKK466 pKa = 10.53QFNDD470 pKa = 3.67DD471 pKa = 3.34ATAGPLLRR479 pKa = 11.84AFGVKK484 pKa = 9.83GKK486 pKa = 10.4YY487 pKa = 8.78GLKK490 pKa = 10.22AAIEE494 pKa = 4.04QFVWGLYY501 pKa = 10.32DD502 pKa = 3.56RR503 pKa = 11.84VGSEE507 pKa = 4.19DD508 pKa = 3.63LKK510 pKa = 11.0PRR512 pKa = 11.84QLPGLLARR520 pKa = 11.84VGYY523 pKa = 7.67RR524 pKa = 11.84TKK526 pKa = 10.96LLDD529 pKa = 3.36MEE531 pKa = 4.67KK532 pKa = 10.61ALKK535 pKa = 9.94KK536 pKa = 9.72IQAVEE541 pKa = 3.78PLGRR545 pKa = 11.84AVMMLDD551 pKa = 3.04ATEE554 pKa = 4.19QCFSSPLFNAISEE567 pKa = 4.55AVTEE571 pKa = 4.06LHH573 pKa = 6.54SNPRR577 pKa = 11.84SGWRR581 pKa = 11.84NYY583 pKa = 9.62LVRR586 pKa = 11.84ASSAWADD593 pKa = 3.14LWDD596 pKa = 3.9EE597 pKa = 4.57LKK599 pKa = 10.87SCGTIVEE606 pKa = 4.5LDD608 pKa = 3.02WSKK611 pKa = 11.06FDD613 pKa = 4.01RR614 pKa = 11.84DD615 pKa = 3.77RR616 pKa = 11.84PAEE619 pKa = 4.6DD620 pKa = 2.75IQFFVDD626 pKa = 4.62VIISCFRR633 pKa = 11.84PKK635 pKa = 10.44NSRR638 pKa = 11.84EE639 pKa = 3.63RR640 pKa = 11.84RR641 pKa = 11.84LLAGYY646 pKa = 10.27RR647 pKa = 11.84KK648 pKa = 9.2MMEE651 pKa = 3.77NALVHH656 pKa = 5.72RR657 pKa = 11.84VMMLDD662 pKa = 3.75DD663 pKa = 4.64GSFFTLEE670 pKa = 3.86GMVPSGSLWTGICDD684 pKa = 3.35TALNIMYY691 pKa = 8.2ITSALRR697 pKa = 11.84KK698 pKa = 9.97LGFDD702 pKa = 4.65DD703 pKa = 4.07STFSPKK709 pKa = 10.26CAGDD713 pKa = 4.26DD714 pKa = 4.03NLTCFQRR721 pKa = 11.84RR722 pKa = 11.84QGLDD726 pKa = 2.88VMEE729 pKa = 4.78RR730 pKa = 11.84LRR732 pKa = 11.84TTLNEE737 pKa = 3.85MFRR740 pKa = 11.84ANIDD744 pKa = 3.3KK745 pKa = 10.77KK746 pKa = 10.94DD747 pKa = 4.53FIVHH751 pKa = 5.56YY752 pKa = 9.38PPYY755 pKa = 10.94AVTTVQACFEE765 pKa = 4.93PGTDD769 pKa = 3.55LSKK772 pKa = 10.44GTSRR776 pKa = 11.84MMDD779 pKa = 3.04KK780 pKa = 11.24AEE782 pKa = 3.99WVPFTGPCPIDD793 pKa = 3.34QAAGRR798 pKa = 11.84SHH800 pKa = 6.25RR801 pKa = 11.84WKK803 pKa = 10.83YY804 pKa = 11.29VFTNKK809 pKa = 9.86PKK811 pKa = 10.24FLANFFLPDD820 pKa = 3.91GKK822 pKa = 9.73PIRR825 pKa = 11.84PAHH828 pKa = 6.75DD829 pKa = 3.76NLEE832 pKa = 4.15KK833 pKa = 10.93LLWPEE838 pKa = 5.16GIHH841 pKa = 6.89EE842 pKa = 4.64SIEE845 pKa = 4.25DD846 pKa = 3.39YY847 pKa = 10.22MAAVLSMVVDD857 pKa = 3.74NPFNHH862 pKa = 6.68HH863 pKa = 5.94NVNHH867 pKa = 5.5MMHH870 pKa = 7.36RR871 pKa = 11.84YY872 pKa = 9.94LIAQQIQKK880 pKa = 10.24QSIFIDD886 pKa = 3.58PAVVMEE892 pKa = 4.45LAKK895 pKa = 10.01TRR897 pKa = 11.84PKK899 pKa = 10.47HH900 pKa = 5.75GEE902 pKa = 3.61AVPFPEE908 pKa = 3.16IAYY911 pKa = 9.24YY912 pKa = 9.86RR913 pKa = 11.84KK914 pKa = 10.02CEE916 pKa = 4.11GYY918 pKa = 10.72VDD920 pKa = 3.9IEE922 pKa = 4.31RR923 pKa = 11.84EE924 pKa = 3.99PEE926 pKa = 3.67FQEE929 pKa = 4.12FFKK932 pKa = 11.02SFRR935 pKa = 11.84EE936 pKa = 4.57FISSVSTLYY945 pKa = 10.78ARR947 pKa = 11.84RR948 pKa = 11.84SEE950 pKa = 4.13GGIDD954 pKa = 2.83AWRR957 pKa = 11.84FMDD960 pKa = 5.13IIRR963 pKa = 11.84GQHH966 pKa = 6.21SIGAGQFGNSVHH978 pKa = 5.32EE979 pKa = 4.07WCRR982 pKa = 11.84FLGEE986 pKa = 4.36HH987 pKa = 6.9PLTRR991 pKa = 11.84SLRR994 pKa = 11.84KK995 pKa = 9.44ARR997 pKa = 11.84RR998 pKa = 11.84FRR1000 pKa = 11.84PMKK1003 pKa = 9.28EE1004 pKa = 3.64AVVADD1009 pKa = 4.24PEE1011 pKa = 4.49TLGKK1015 pKa = 9.95VEE1017 pKa = 5.52KK1018 pKa = 10.85AFTWVLDD1025 pKa = 3.63LCEE1028 pKa = 5.48ANDD1031 pKa = 3.56PMTPLFYY1038 pKa = 10.85ASAVSDD1044 pKa = 3.36ILSSNVSAPP1053 pKa = 3.23
Molecular weight: 118.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1438 |
385 |
1053 |
719.0 |
80.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.11 ± 0.887 | 1.599 ± 0.148 |
6.745 ± 0.644 | 5.981 ± 0.253 |
3.894 ± 0.639 | 6.05 ± 0.293 |
2.225 ± 0.055 | 4.242 ± 0.554 |
6.537 ± 0.619 | 10.083 ± 0.361 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.99 ± 0.45 | 3.06 ± 0.028 |
6.05 ± 0.602 | 4.659 ± 0.648 |
6.815 ± 0.414 | 5.981 ± 0.131 |
4.798 ± 0.451 | 5.633 ± 0.472 |
1.46 ± 0.336 | 2.086 ± 0.388 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
