
Nanosalinarum sp. (strain J07AB56)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Nanohaloarchaea; Candidatus Nanosalinarum; unclassified Candidatus Nanosalinarum
Average proteome isoelectric point is 5.15
Get precalculated fractions of proteins
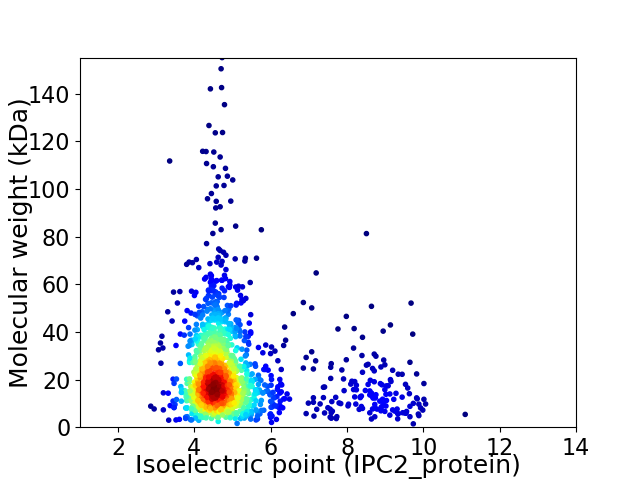
Virtual 2D-PAGE plot for 1402 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G0QCW2|G0QCW2_NANSJ Small-conductance mechanosensitive channel OS=Nanosalinarum sp. (strain J07AB56) OX=889962 GN=J07AB56_08380 PE=3 SV=1
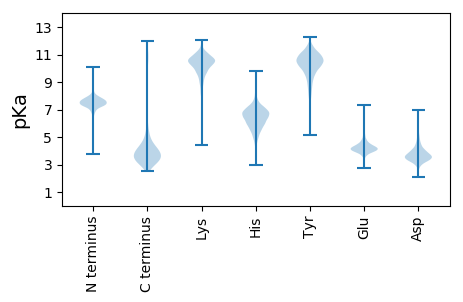
DDD2 pKa = 6.29DD3 pKa = 4.72SDDD6 pKa = 3.94KK7 pKa = 11.04YY8 pKa = 10.54QGSFGSTTSPVPNSWDDD25 pKa = 3.33RR26 pKa = 11.84STGFLGLFGAQEEE39 pKa = 4.37YY40 pKa = 10.67DDD42 pKa = 4.33PRR44 pKa = 11.84GDDD47 pKa = 3.52DD48 pKa = 5.19GDDD51 pKa = 3.58DDD53 pKa = 5.97DD54 pKa = 5.53DD55 pKa = 5.65EEE57 pKa = 4.56TLDDD61 pKa = 3.58PTDDD65 pKa = 3.05GMAEEE70 pKa = 4.71DD71 pKa = 4.58GSSDDD76 pKa = 3.49SVSQIEEE83 pKa = 4.62KKK85 pKa = 9.7EEE87 pKa = 3.94GTPNANNNIRR97 pKa = 11.84IEEE100 pKa = 4.12EE101 pKa = 4.24EEE103 pKa = 4.3TKKK106 pKa = 10.32CDDD109 pKa = 3.29DDD111 pKa = 4.59GSCPSISSNFLPTYYY126 pKa = 9.84GGPNYYY132 pKa = 10.39IAQSNSDDD140 pKa = 3.55LKKK143 pKa = 10.03EE144 pKa = 3.64SKKK147 pKa = 9.69SRR149 pKa = 11.84GTTTLGSTTDDD160 pKa = 3.28YY161 pKa = 11.25DD162 pKa = 4.03EEE164 pKa = 4.18YY165 pKa = 10.84SYYY168 pKa = 11.25VVQSTGSATDDD179 pKa = 3.28SCTATGNYYY188 pKa = 9.37CNLDDD193 pKa = 3.3SNTVTTSKKK202 pKa = 9.95EEE204 pKa = 4.08SHHH207 pKa = 7.35DD208 pKa = 4.11YYY210 pKa = 11.33LQPDDD215 pKa = 4.17EE216 pKa = 5.5DD217 pKa = 4.09HHH219 pKa = 6.88ATTSDDD225 pKa = 4.91EEE227 pKa = 4.44TSNGVGYYY235 pKa = 10.43LNEEE239 pKa = 3.81TDDD242 pKa = 3.58LRR244 pKa = 11.84LEEE247 pKa = 4.36AHHH250 pKa = 7.55DD251 pKa = 3.72DD252 pKa = 4.97FDDD255 pKa = 3.49GNDDD259 pKa = 3.86IEEE262 pKa = 4.55EE263 pKa = 4.39PGGSTTIRR271 pKa = 11.84SYYY274 pKa = 11.32CSLDDD279 pKa = 3.31TPKKK283 pKa = 10.75DD284 pKa = 3.82SCDDD288 pKa = 3.18SNTGTYYY295 pKa = 7.16TYYY298 pKa = 11.0FTTSSPKKK306 pKa = 9.0TGTTSSQEEE315 pKa = 3.74FRR317 pKa = 11.84YYY319 pKa = 9.96EE320 pKa = 4.06VPSSGWPKKK329 pKa = 9.99TVLDDD334 pKa = 4.23NNDDD338 pKa = 2.98TGAVGEEE345 pKa = 4.51DD346 pKa = 4.06YY347 pKa = 10.03TVHHH351 pKa = 6.61HH352 pKa = 6.8KK353 pKa = 10.95KK354 pKa = 10.48EE355 pKa = 4.07GTGYYY360 pKa = 10.59SVDDD364 pKa = 3.31GRR366 pKa = 11.84DDD368 pKa = 3.19VDDD371 pKa = 3.75GNNRR375 pKa = 11.84AFGWSTFDDD384 pKa = 3.18TMDDD388 pKa = 3.67DDD390 pKa = 3.9EE391 pKa = 4.68ATEEE395 pKa = 4.18TGSEEE400 pKa = 4.05YYY402 pKa = 10.09EE403 pKa = 3.75DDD405 pKa = 3.57NNGEEE410 pKa = 4.29DDD412 pKa = 3.7LVAVHHH418 pKa = 6.47EE419 pKa = 4.54EEE421 pKa = 4.54VGDDD425 pKa = 3.68SNFGTYYY432 pKa = 10.52SRR434 pKa = 11.84SDDD437 pKa = 4.33SVSCPGDDD445 pKa = 3.35YY446 pKa = 8.99KKK448 pKa = 10.87VAAVDDD454 pKa = 3.95SLNSFD
DDD2 pKa = 6.29DD3 pKa = 4.72SDDD6 pKa = 3.94KK7 pKa = 11.04YY8 pKa = 10.54QGSFGSTTSPVPNSWDDD25 pKa = 3.33RR26 pKa = 11.84STGFLGLFGAQEEE39 pKa = 4.37YY40 pKa = 10.67DDD42 pKa = 4.33PRR44 pKa = 11.84GDDD47 pKa = 3.52DD48 pKa = 5.19GDDD51 pKa = 3.58DDD53 pKa = 5.97DD54 pKa = 5.53DD55 pKa = 5.65EEE57 pKa = 4.56TLDDD61 pKa = 3.58PTDDD65 pKa = 3.05GMAEEE70 pKa = 4.71DD71 pKa = 4.58GSSDDD76 pKa = 3.49SVSQIEEE83 pKa = 4.62KKK85 pKa = 9.7EEE87 pKa = 3.94GTPNANNNIRR97 pKa = 11.84IEEE100 pKa = 4.12EE101 pKa = 4.24EEE103 pKa = 4.3TKKK106 pKa = 10.32CDDD109 pKa = 3.29DDD111 pKa = 4.59GSCPSISSNFLPTYYY126 pKa = 9.84GGPNYYY132 pKa = 10.39IAQSNSDDD140 pKa = 3.55LKKK143 pKa = 10.03EE144 pKa = 3.64SKKK147 pKa = 9.69SRR149 pKa = 11.84GTTTLGSTTDDD160 pKa = 3.28YY161 pKa = 11.25DD162 pKa = 4.03EEE164 pKa = 4.18YY165 pKa = 10.84SYYY168 pKa = 11.25VVQSTGSATDDD179 pKa = 3.28SCTATGNYYY188 pKa = 9.37CNLDDD193 pKa = 3.3SNTVTTSKKK202 pKa = 9.95EEE204 pKa = 4.08SHHH207 pKa = 7.35DD208 pKa = 4.11YYY210 pKa = 11.33LQPDDD215 pKa = 4.17EE216 pKa = 5.5DD217 pKa = 4.09HHH219 pKa = 6.88ATTSDDD225 pKa = 4.91EEE227 pKa = 4.44TSNGVGYYY235 pKa = 10.43LNEEE239 pKa = 3.81TDDD242 pKa = 3.58LRR244 pKa = 11.84LEEE247 pKa = 4.36AHHH250 pKa = 7.55DD251 pKa = 3.72DD252 pKa = 4.97FDDD255 pKa = 3.49GNDDD259 pKa = 3.86IEEE262 pKa = 4.55EE263 pKa = 4.39PGGSTTIRR271 pKa = 11.84SYYY274 pKa = 11.32CSLDDD279 pKa = 3.31TPKKK283 pKa = 10.75DD284 pKa = 3.82SCDDD288 pKa = 3.18SNTGTYYY295 pKa = 7.16TYYY298 pKa = 11.0FTTSSPKKK306 pKa = 9.0TGTTSSQEEE315 pKa = 3.74FRR317 pKa = 11.84YYY319 pKa = 9.96EE320 pKa = 4.06VPSSGWPKKK329 pKa = 9.99TVLDDD334 pKa = 4.23NNDDD338 pKa = 2.98TGAVGEEE345 pKa = 4.51DD346 pKa = 4.06YY347 pKa = 10.03TVHHH351 pKa = 6.61HH352 pKa = 6.8KK353 pKa = 10.95KK354 pKa = 10.48EE355 pKa = 4.07GTGYYY360 pKa = 10.59SVDDD364 pKa = 3.31GRR366 pKa = 11.84DDD368 pKa = 3.19VDDD371 pKa = 3.75GNNRR375 pKa = 11.84AFGWSTFDDD384 pKa = 3.18TMDDD388 pKa = 3.67DDD390 pKa = 3.9EE391 pKa = 4.68ATEEE395 pKa = 4.18TGSEEE400 pKa = 4.05YYY402 pKa = 10.09EE403 pKa = 3.75DDD405 pKa = 3.57NNGEEE410 pKa = 4.29DDD412 pKa = 3.7LVAVHHH418 pKa = 6.47EE419 pKa = 4.54EEE421 pKa = 4.54VGDDD425 pKa = 3.68SNFGTYYY432 pKa = 10.52SRR434 pKa = 11.84SDDD437 pKa = 4.33SVSCPGDDD445 pKa = 3.35YY446 pKa = 8.99KKK448 pKa = 10.87VAAVDDD454 pKa = 3.95SLNSFD
Molecular weight: 48.91 kDa
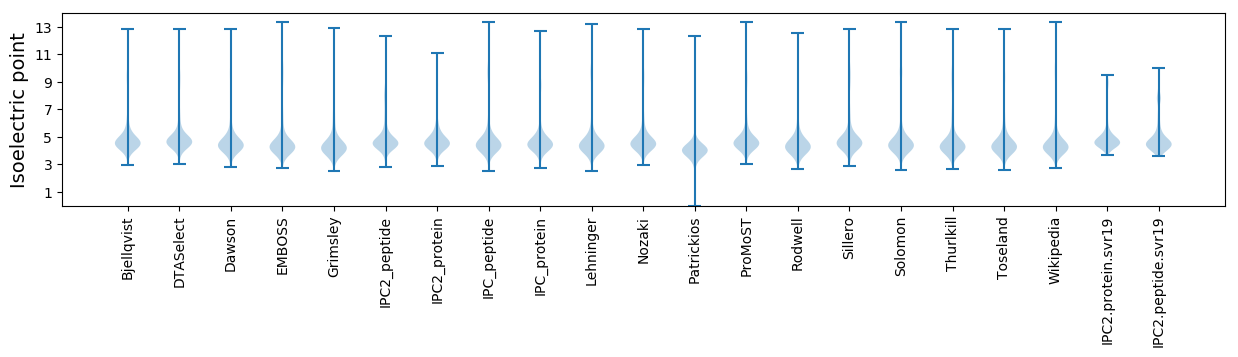
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G0QDK0|G0QDK0_NANSJ DNA-binding ferritin-like protein oxidative damage protectant OS=Nanosalinarum sp. (strain J07AB56) OX=889962 GN=J07AB56_10770 PE=3 SV=1
MM1 pKa = 7.56RR2 pKa = 11.84QLKK5 pKa = 10.26LGSKK9 pKa = 9.4HH10 pKa = 5.87SRR12 pKa = 11.84RR13 pKa = 11.84LTHH16 pKa = 5.57QTPKK20 pKa = 9.28MKK22 pKa = 10.18TMRR25 pKa = 11.84TMSLPLSRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84SWKK39 pKa = 9.08VHH41 pKa = 4.18VRR43 pKa = 11.84HH44 pKa = 6.31
MM1 pKa = 7.56RR2 pKa = 11.84QLKK5 pKa = 10.26LGSKK9 pKa = 9.4HH10 pKa = 5.87SRR12 pKa = 11.84RR13 pKa = 11.84LTHH16 pKa = 5.57QTPKK20 pKa = 9.28MKK22 pKa = 10.18TMRR25 pKa = 11.84TMSLPLSRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84SWKK39 pKa = 9.08VHH41 pKa = 4.18VRR43 pKa = 11.84HH44 pKa = 6.31
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
319963 |
13 |
1377 |
228.2 |
25.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.615 ± 0.08 | 0.711 ± 0.02 |
7.512 ± 0.073 | 9.843 ± 0.115 |
3.515 ± 0.048 | 7.875 ± 0.076 |
1.803 ± 0.037 | 4.376 ± 0.051 |
3.695 ± 0.059 | 8.79 ± 0.091 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.127 ± 0.034 | 3.361 ± 0.051 |
3.869 ± 0.043 | 3.577 ± 0.048 |
6.368 ± 0.07 | 7.575 ± 0.077 |
5.561 ± 0.06 | 8.051 ± 0.076 |
0.854 ± 0.025 | 2.701 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
