
Pleurocapsa sp. PCC 7327
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Pleurocapsales; Hyellaceae; Pleurocapsa; unclassified Pleurocapsa
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
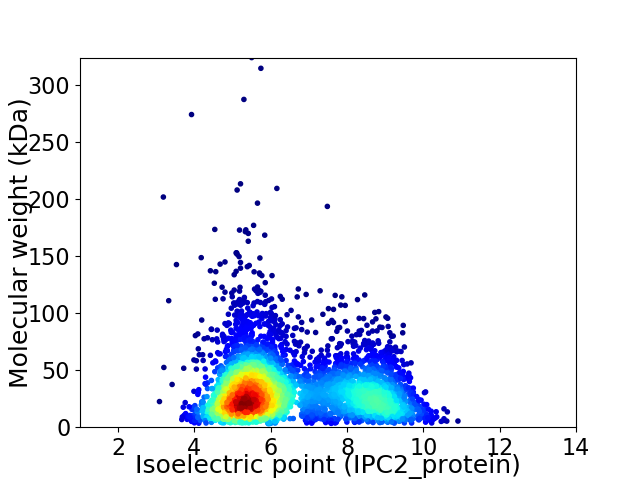
Virtual 2D-PAGE plot for 4255 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9TB87|K9TB87_9CYAN Ferredoxin (2Fe-2S) OS=Pleurocapsa sp. PCC 7327 OX=118163 GN=Ple7327_4593 PE=3 SV=1
MM1 pKa = 7.76ANNNDD6 pKa = 4.2IIQLTNNNFDD16 pKa = 4.11DD17 pKa = 4.17YY18 pKa = 11.72GPRR21 pKa = 11.84IDD23 pKa = 3.83GNKK26 pKa = 9.32VVWTGYY32 pKa = 10.01PDD34 pKa = 4.33FNDD37 pKa = 3.64SEE39 pKa = 4.76IYY41 pKa = 10.41FYY43 pKa = 11.03DD44 pKa = 3.92GSKK47 pKa = 8.59ITQLTNNNFYY57 pKa = 10.86DD58 pKa = 3.89YY59 pKa = 11.18GPQISGNNVVWSGSDD74 pKa = 3.47GNDD77 pKa = 3.5DD78 pKa = 4.99EE79 pKa = 6.4IYY81 pKa = 10.57LYY83 pKa = 10.84DD84 pKa = 3.83GNKK87 pKa = 6.51TTRR90 pKa = 11.84LTDD93 pKa = 3.3NNLYY97 pKa = 10.54DD98 pKa = 4.23YY99 pKa = 11.17SPQIDD104 pKa = 3.82GNKK107 pKa = 8.47VVWYY111 pKa = 9.0AYY113 pKa = 9.66PPDD116 pKa = 3.96YY117 pKa = 10.62SDD119 pKa = 4.1SEE121 pKa = 4.38IYY123 pKa = 10.4FYY125 pKa = 11.12DD126 pKa = 3.75GSKK129 pKa = 9.97IVQLTDD135 pKa = 3.37NNFSDD140 pKa = 4.57YY141 pKa = 11.22SPQISGNKK149 pKa = 8.47VVWSGYY155 pKa = 10.33DD156 pKa = 3.37GNDD159 pKa = 3.52DD160 pKa = 4.95EE161 pKa = 6.35IYY163 pKa = 10.54LYY165 pKa = 10.83DD166 pKa = 3.86GSKK169 pKa = 9.7TIQLTNNNTYY179 pKa = 10.55DD180 pKa = 3.75YY181 pKa = 10.84SPQIDD186 pKa = 3.65GDD188 pKa = 4.02KK189 pKa = 10.78VVWSGYY195 pKa = 10.35DD196 pKa = 3.37GNDD199 pKa = 3.52DD200 pKa = 4.95EE201 pKa = 6.35IYY203 pKa = 10.5LYY205 pKa = 10.74DD206 pKa = 3.72GSKK209 pKa = 9.94IVQLTDD215 pKa = 3.37NNFSDD220 pKa = 4.57YY221 pKa = 11.22SPQISGNKK229 pKa = 8.41VVWSGYY235 pKa = 9.67PDD237 pKa = 4.01FNDD240 pKa = 3.82SEE242 pKa = 4.59IYY244 pKa = 10.53LYY246 pKa = 10.89DD247 pKa = 3.91GNQTIQLTDD256 pKa = 3.33NNFYY260 pKa = 10.76DD261 pKa = 4.2YY262 pKa = 11.16SPQISGDD269 pKa = 3.36KK270 pKa = 9.7VVWYY274 pKa = 8.7GYY276 pKa = 9.48PDD278 pKa = 3.52NADD281 pKa = 3.57SEE283 pKa = 4.91IYY285 pKa = 10.37FYY287 pKa = 11.3DD288 pKa = 4.3GDD290 pKa = 4.59VIQLTNNNTYY300 pKa = 10.72DD301 pKa = 4.0YY302 pKa = 10.95DD303 pKa = 3.95PQISGNKK310 pKa = 8.44VVWSSYY316 pKa = 11.06DD317 pKa = 3.23GTDD320 pKa = 3.16RR321 pKa = 11.84EE322 pKa = 4.34IYY324 pKa = 10.19LYY326 pKa = 9.25EE327 pKa = 4.34IPPGIEE333 pKa = 4.24LIGTEE338 pKa = 4.31GDD340 pKa = 3.81DD341 pKa = 3.42VLTGSRR347 pKa = 11.84GPDD350 pKa = 3.4TISGLGGNDD359 pKa = 3.18VLQGLAGNDD368 pKa = 3.54QILGGSGNDD377 pKa = 4.67LITAGDD383 pKa = 4.0GKK385 pKa = 10.74DD386 pKa = 3.27KK387 pKa = 11.74AEE389 pKa = 4.47GGSGNDD395 pKa = 3.65TISGNGGNDD404 pKa = 2.78ILIGNEE410 pKa = 3.61GRR412 pKa = 11.84DD413 pKa = 4.0DD414 pKa = 3.68ILGGEE419 pKa = 4.76DD420 pKa = 3.69NDD422 pKa = 5.63SIDD425 pKa = 3.53GGAGRR430 pKa = 11.84DD431 pKa = 3.48RR432 pKa = 11.84LLGQAGNDD440 pKa = 3.83TIVGQGGDD448 pKa = 3.67DD449 pKa = 4.08TIDD452 pKa = 3.83GGDD455 pKa = 3.79GTDD458 pKa = 3.59SLSGNAGSDD467 pKa = 3.64RR468 pKa = 11.84ILGGSGSDD476 pKa = 3.4SLSGGTEE483 pKa = 3.69ADD485 pKa = 3.69TLVGGFDD492 pKa = 3.84NDD494 pKa = 4.13SLDD497 pKa = 4.04GGAGNDD503 pKa = 3.36NLVGINLAVVSGAKK517 pKa = 10.03VGFGAGEE524 pKa = 4.26IDD526 pKa = 3.91TLSGGTGYY534 pKa = 8.56DD535 pKa = 3.34TFALGDD541 pKa = 3.87NDD543 pKa = 3.75RR544 pKa = 11.84VYY546 pKa = 11.48YY547 pKa = 10.56DD548 pKa = 5.07DD549 pKa = 5.94GDD551 pKa = 4.06PLTTGEE557 pKa = 4.48SDD559 pKa = 3.37YY560 pKa = 11.7TLITDD565 pKa = 4.29YY566 pKa = 11.4DD567 pKa = 3.96PSQDD571 pKa = 3.12SLQRR575 pKa = 11.84KK576 pKa = 9.37GSDD579 pKa = 3.18DD580 pKa = 4.28LYY582 pKa = 11.26KK583 pKa = 10.44LDD585 pKa = 4.43YY586 pKa = 8.73FTTPSGRR593 pKa = 11.84VDD595 pKa = 2.89VAMIYY600 pKa = 10.67DD601 pKa = 4.03PGVEE605 pKa = 4.14ARR607 pKa = 11.84GEE609 pKa = 4.27TIAILQGSGASSASTASLSAQQTTDD634 pKa = 3.2NTDD637 pKa = 3.06EE638 pKa = 4.42SPIPEE643 pKa = 4.24NTSPLARR650 pKa = 11.84LAGLAEE656 pKa = 4.42GKK658 pKa = 10.2DD659 pKa = 3.86YY660 pKa = 11.5ASGEE664 pKa = 4.41LIVKK668 pKa = 9.57LKK670 pKa = 10.97AGTEE674 pKa = 4.01ANKK677 pKa = 10.13ISLLQEE683 pKa = 3.75RR684 pKa = 11.84LGATVLEE691 pKa = 4.52TTKK694 pKa = 10.55TLGIQRR700 pKa = 11.84WKK702 pKa = 10.79LEE704 pKa = 4.01EE705 pKa = 4.28GTSVRR710 pKa = 11.84DD711 pKa = 4.19AIASFSADD719 pKa = 3.3PAIEE723 pKa = 4.3YY724 pKa = 10.2IEE726 pKa = 4.35PNYY729 pKa = 9.97IRR731 pKa = 11.84STTATIPNDD740 pKa = 3.34PRR742 pKa = 11.84FSEE745 pKa = 4.41LWGLNNTGQTGGTPDD760 pKa = 4.04ADD762 pKa = 3.22IDD764 pKa = 4.82APEE767 pKa = 3.94AWDD770 pKa = 3.77RR771 pKa = 11.84QTGSDD776 pKa = 3.76VVVGVIDD783 pKa = 3.44TGVDD787 pKa = 3.57YY788 pKa = 11.17NHH790 pKa = 7.81PDD792 pKa = 3.46LNDD795 pKa = 3.71NMWTNPGEE803 pKa = 4.38TPNNGVDD810 pKa = 3.78DD811 pKa = 5.12DD812 pKa = 5.07GNGYY816 pKa = 9.87VDD818 pKa = 5.48DD819 pKa = 4.46YY820 pKa = 11.47YY821 pKa = 11.58GYY823 pKa = 11.24DD824 pKa = 3.58FVNEE828 pKa = 4.7DD829 pKa = 4.25GDD831 pKa = 3.95PFDD834 pKa = 5.8DD835 pKa = 4.74FFHH838 pKa = 6.28GTHH841 pKa = 5.55VAGTIAAEE849 pKa = 4.44GNNNTGVTGVNWNAKK864 pKa = 8.84IMAIKK869 pKa = 10.23FLDD872 pKa = 3.84SGGSGTDD879 pKa = 3.45FDD881 pKa = 5.82AIEE884 pKa = 4.08AVEE887 pKa = 4.14YY888 pKa = 8.63STLMGVKK895 pKa = 8.92LTSNSWGGYY904 pKa = 8.3FFSQGLYY911 pKa = 10.56DD912 pKa = 5.52AIAAAGKK919 pKa = 10.13AGQLFIAAAGNGTNDD934 pKa = 3.36NDD936 pKa = 4.6GPNPAYY942 pKa = 9.09PASYY946 pKa = 10.81DD947 pKa = 3.45LDD949 pKa = 4.46NIISVAATDD958 pKa = 3.95ASDD961 pKa = 3.39RR962 pKa = 11.84LAWFSNYY969 pKa = 9.6GATSVDD975 pKa = 3.35LGAPGVSVLSTVPGGGYY992 pKa = 10.5ASYY995 pKa = 10.79DD996 pKa = 3.87GTSMATPHH1004 pKa = 5.64VSGVASLLWSEE1015 pKa = 4.97DD1016 pKa = 3.68PNLSAQEE1023 pKa = 4.12VKK1025 pKa = 10.66EE1026 pKa = 4.03LLLTTVDD1033 pKa = 3.84PLTVLEE1039 pKa = 4.65GKK1041 pKa = 7.87TVSGGRR1047 pKa = 11.84LNAFNALSEE1056 pKa = 4.38LGPPNEE1062 pKa = 4.61IIGTEE1067 pKa = 4.06GDD1069 pKa = 3.89DD1070 pKa = 4.79VITGTNRR1077 pKa = 11.84RR1078 pKa = 11.84DD1079 pKa = 3.64RR1080 pKa = 11.84ISGLGGNDD1088 pKa = 3.51IIQALAGRR1096 pKa = 11.84DD1097 pKa = 3.76EE1098 pKa = 4.39IFGGDD1103 pKa = 3.72GDD1105 pKa = 4.84DD1106 pKa = 6.49LITADD1111 pKa = 4.18EE1112 pKa = 4.76GNDD1115 pKa = 3.67TVEE1118 pKa = 4.5GGAGRR1123 pKa = 11.84DD1124 pKa = 3.21RR1125 pKa = 11.84IFGNDD1130 pKa = 3.43GDD1132 pKa = 5.53DD1133 pKa = 3.6LLKK1136 pKa = 11.4GNGGRR1141 pKa = 11.84DD1142 pKa = 3.51NILGGDD1148 pKa = 3.88GNDD1151 pKa = 4.09SIEE1154 pKa = 4.34GGAGNDD1160 pKa = 3.24RR1161 pKa = 11.84LLGEE1165 pKa = 4.89SGDD1168 pKa = 3.91DD1169 pKa = 3.8TILGNAGQDD1178 pKa = 3.38TLDD1181 pKa = 3.78GGEE1184 pKa = 4.91GKK1186 pKa = 10.29DD1187 pKa = 3.68SLNGGTDD1194 pKa = 2.93NDD1196 pKa = 5.24LIFGGKK1202 pKa = 10.34GEE1204 pKa = 4.46DD1205 pKa = 3.67SLVGEE1210 pKa = 5.13AGADD1214 pKa = 3.61SLTGGASNDD1223 pKa = 3.89SLDD1226 pKa = 3.93GGAGSDD1232 pKa = 3.4RR1233 pKa = 11.84LVGVDD1238 pKa = 3.91PSASYY1243 pKa = 10.77GAGEE1247 pKa = 4.65LDD1249 pKa = 3.76VLTGFSDD1256 pKa = 3.19SDD1258 pKa = 3.85TFVLGDD1264 pKa = 3.79ASRR1267 pKa = 11.84VFYY1270 pKa = 11.11SDD1272 pKa = 4.28GDD1274 pKa = 3.9PLTTGEE1280 pKa = 4.45SDD1282 pKa = 3.62YY1283 pKa = 12.0ALISDD1288 pKa = 4.97FDD1290 pKa = 4.16SSQDD1294 pKa = 3.23IIQLHH1299 pKa = 5.7GAIAQYY1305 pKa = 11.26DD1306 pKa = 4.0LDD1308 pKa = 4.48FFTTSSGSINAALIYY1323 pKa = 10.67DD1324 pKa = 4.38PGDD1327 pKa = 3.33AARR1330 pKa = 11.84GEE1332 pKa = 4.73LISILQNVPTDD1343 pKa = 4.12LNLGSPAFSFVV1354 pKa = 2.94
MM1 pKa = 7.76ANNNDD6 pKa = 4.2IIQLTNNNFDD16 pKa = 4.11DD17 pKa = 4.17YY18 pKa = 11.72GPRR21 pKa = 11.84IDD23 pKa = 3.83GNKK26 pKa = 9.32VVWTGYY32 pKa = 10.01PDD34 pKa = 4.33FNDD37 pKa = 3.64SEE39 pKa = 4.76IYY41 pKa = 10.41FYY43 pKa = 11.03DD44 pKa = 3.92GSKK47 pKa = 8.59ITQLTNNNFYY57 pKa = 10.86DD58 pKa = 3.89YY59 pKa = 11.18GPQISGNNVVWSGSDD74 pKa = 3.47GNDD77 pKa = 3.5DD78 pKa = 4.99EE79 pKa = 6.4IYY81 pKa = 10.57LYY83 pKa = 10.84DD84 pKa = 3.83GNKK87 pKa = 6.51TTRR90 pKa = 11.84LTDD93 pKa = 3.3NNLYY97 pKa = 10.54DD98 pKa = 4.23YY99 pKa = 11.17SPQIDD104 pKa = 3.82GNKK107 pKa = 8.47VVWYY111 pKa = 9.0AYY113 pKa = 9.66PPDD116 pKa = 3.96YY117 pKa = 10.62SDD119 pKa = 4.1SEE121 pKa = 4.38IYY123 pKa = 10.4FYY125 pKa = 11.12DD126 pKa = 3.75GSKK129 pKa = 9.97IVQLTDD135 pKa = 3.37NNFSDD140 pKa = 4.57YY141 pKa = 11.22SPQISGNKK149 pKa = 8.47VVWSGYY155 pKa = 10.33DD156 pKa = 3.37GNDD159 pKa = 3.52DD160 pKa = 4.95EE161 pKa = 6.35IYY163 pKa = 10.54LYY165 pKa = 10.83DD166 pKa = 3.86GSKK169 pKa = 9.7TIQLTNNNTYY179 pKa = 10.55DD180 pKa = 3.75YY181 pKa = 10.84SPQIDD186 pKa = 3.65GDD188 pKa = 4.02KK189 pKa = 10.78VVWSGYY195 pKa = 10.35DD196 pKa = 3.37GNDD199 pKa = 3.52DD200 pKa = 4.95EE201 pKa = 6.35IYY203 pKa = 10.5LYY205 pKa = 10.74DD206 pKa = 3.72GSKK209 pKa = 9.94IVQLTDD215 pKa = 3.37NNFSDD220 pKa = 4.57YY221 pKa = 11.22SPQISGNKK229 pKa = 8.41VVWSGYY235 pKa = 9.67PDD237 pKa = 4.01FNDD240 pKa = 3.82SEE242 pKa = 4.59IYY244 pKa = 10.53LYY246 pKa = 10.89DD247 pKa = 3.91GNQTIQLTDD256 pKa = 3.33NNFYY260 pKa = 10.76DD261 pKa = 4.2YY262 pKa = 11.16SPQISGDD269 pKa = 3.36KK270 pKa = 9.7VVWYY274 pKa = 8.7GYY276 pKa = 9.48PDD278 pKa = 3.52NADD281 pKa = 3.57SEE283 pKa = 4.91IYY285 pKa = 10.37FYY287 pKa = 11.3DD288 pKa = 4.3GDD290 pKa = 4.59VIQLTNNNTYY300 pKa = 10.72DD301 pKa = 4.0YY302 pKa = 10.95DD303 pKa = 3.95PQISGNKK310 pKa = 8.44VVWSSYY316 pKa = 11.06DD317 pKa = 3.23GTDD320 pKa = 3.16RR321 pKa = 11.84EE322 pKa = 4.34IYY324 pKa = 10.19LYY326 pKa = 9.25EE327 pKa = 4.34IPPGIEE333 pKa = 4.24LIGTEE338 pKa = 4.31GDD340 pKa = 3.81DD341 pKa = 3.42VLTGSRR347 pKa = 11.84GPDD350 pKa = 3.4TISGLGGNDD359 pKa = 3.18VLQGLAGNDD368 pKa = 3.54QILGGSGNDD377 pKa = 4.67LITAGDD383 pKa = 4.0GKK385 pKa = 10.74DD386 pKa = 3.27KK387 pKa = 11.74AEE389 pKa = 4.47GGSGNDD395 pKa = 3.65TISGNGGNDD404 pKa = 2.78ILIGNEE410 pKa = 3.61GRR412 pKa = 11.84DD413 pKa = 4.0DD414 pKa = 3.68ILGGEE419 pKa = 4.76DD420 pKa = 3.69NDD422 pKa = 5.63SIDD425 pKa = 3.53GGAGRR430 pKa = 11.84DD431 pKa = 3.48RR432 pKa = 11.84LLGQAGNDD440 pKa = 3.83TIVGQGGDD448 pKa = 3.67DD449 pKa = 4.08TIDD452 pKa = 3.83GGDD455 pKa = 3.79GTDD458 pKa = 3.59SLSGNAGSDD467 pKa = 3.64RR468 pKa = 11.84ILGGSGSDD476 pKa = 3.4SLSGGTEE483 pKa = 3.69ADD485 pKa = 3.69TLVGGFDD492 pKa = 3.84NDD494 pKa = 4.13SLDD497 pKa = 4.04GGAGNDD503 pKa = 3.36NLVGINLAVVSGAKK517 pKa = 10.03VGFGAGEE524 pKa = 4.26IDD526 pKa = 3.91TLSGGTGYY534 pKa = 8.56DD535 pKa = 3.34TFALGDD541 pKa = 3.87NDD543 pKa = 3.75RR544 pKa = 11.84VYY546 pKa = 11.48YY547 pKa = 10.56DD548 pKa = 5.07DD549 pKa = 5.94GDD551 pKa = 4.06PLTTGEE557 pKa = 4.48SDD559 pKa = 3.37YY560 pKa = 11.7TLITDD565 pKa = 4.29YY566 pKa = 11.4DD567 pKa = 3.96PSQDD571 pKa = 3.12SLQRR575 pKa = 11.84KK576 pKa = 9.37GSDD579 pKa = 3.18DD580 pKa = 4.28LYY582 pKa = 11.26KK583 pKa = 10.44LDD585 pKa = 4.43YY586 pKa = 8.73FTTPSGRR593 pKa = 11.84VDD595 pKa = 2.89VAMIYY600 pKa = 10.67DD601 pKa = 4.03PGVEE605 pKa = 4.14ARR607 pKa = 11.84GEE609 pKa = 4.27TIAILQGSGASSASTASLSAQQTTDD634 pKa = 3.2NTDD637 pKa = 3.06EE638 pKa = 4.42SPIPEE643 pKa = 4.24NTSPLARR650 pKa = 11.84LAGLAEE656 pKa = 4.42GKK658 pKa = 10.2DD659 pKa = 3.86YY660 pKa = 11.5ASGEE664 pKa = 4.41LIVKK668 pKa = 9.57LKK670 pKa = 10.97AGTEE674 pKa = 4.01ANKK677 pKa = 10.13ISLLQEE683 pKa = 3.75RR684 pKa = 11.84LGATVLEE691 pKa = 4.52TTKK694 pKa = 10.55TLGIQRR700 pKa = 11.84WKK702 pKa = 10.79LEE704 pKa = 4.01EE705 pKa = 4.28GTSVRR710 pKa = 11.84DD711 pKa = 4.19AIASFSADD719 pKa = 3.3PAIEE723 pKa = 4.3YY724 pKa = 10.2IEE726 pKa = 4.35PNYY729 pKa = 9.97IRR731 pKa = 11.84STTATIPNDD740 pKa = 3.34PRR742 pKa = 11.84FSEE745 pKa = 4.41LWGLNNTGQTGGTPDD760 pKa = 4.04ADD762 pKa = 3.22IDD764 pKa = 4.82APEE767 pKa = 3.94AWDD770 pKa = 3.77RR771 pKa = 11.84QTGSDD776 pKa = 3.76VVVGVIDD783 pKa = 3.44TGVDD787 pKa = 3.57YY788 pKa = 11.17NHH790 pKa = 7.81PDD792 pKa = 3.46LNDD795 pKa = 3.71NMWTNPGEE803 pKa = 4.38TPNNGVDD810 pKa = 3.78DD811 pKa = 5.12DD812 pKa = 5.07GNGYY816 pKa = 9.87VDD818 pKa = 5.48DD819 pKa = 4.46YY820 pKa = 11.47YY821 pKa = 11.58GYY823 pKa = 11.24DD824 pKa = 3.58FVNEE828 pKa = 4.7DD829 pKa = 4.25GDD831 pKa = 3.95PFDD834 pKa = 5.8DD835 pKa = 4.74FFHH838 pKa = 6.28GTHH841 pKa = 5.55VAGTIAAEE849 pKa = 4.44GNNNTGVTGVNWNAKK864 pKa = 8.84IMAIKK869 pKa = 10.23FLDD872 pKa = 3.84SGGSGTDD879 pKa = 3.45FDD881 pKa = 5.82AIEE884 pKa = 4.08AVEE887 pKa = 4.14YY888 pKa = 8.63STLMGVKK895 pKa = 8.92LTSNSWGGYY904 pKa = 8.3FFSQGLYY911 pKa = 10.56DD912 pKa = 5.52AIAAAGKK919 pKa = 10.13AGQLFIAAAGNGTNDD934 pKa = 3.36NDD936 pKa = 4.6GPNPAYY942 pKa = 9.09PASYY946 pKa = 10.81DD947 pKa = 3.45LDD949 pKa = 4.46NIISVAATDD958 pKa = 3.95ASDD961 pKa = 3.39RR962 pKa = 11.84LAWFSNYY969 pKa = 9.6GATSVDD975 pKa = 3.35LGAPGVSVLSTVPGGGYY992 pKa = 10.5ASYY995 pKa = 10.79DD996 pKa = 3.87GTSMATPHH1004 pKa = 5.64VSGVASLLWSEE1015 pKa = 4.97DD1016 pKa = 3.68PNLSAQEE1023 pKa = 4.12VKK1025 pKa = 10.66EE1026 pKa = 4.03LLLTTVDD1033 pKa = 3.84PLTVLEE1039 pKa = 4.65GKK1041 pKa = 7.87TVSGGRR1047 pKa = 11.84LNAFNALSEE1056 pKa = 4.38LGPPNEE1062 pKa = 4.61IIGTEE1067 pKa = 4.06GDD1069 pKa = 3.89DD1070 pKa = 4.79VITGTNRR1077 pKa = 11.84RR1078 pKa = 11.84DD1079 pKa = 3.64RR1080 pKa = 11.84ISGLGGNDD1088 pKa = 3.51IIQALAGRR1096 pKa = 11.84DD1097 pKa = 3.76EE1098 pKa = 4.39IFGGDD1103 pKa = 3.72GDD1105 pKa = 4.84DD1106 pKa = 6.49LITADD1111 pKa = 4.18EE1112 pKa = 4.76GNDD1115 pKa = 3.67TVEE1118 pKa = 4.5GGAGRR1123 pKa = 11.84DD1124 pKa = 3.21RR1125 pKa = 11.84IFGNDD1130 pKa = 3.43GDD1132 pKa = 5.53DD1133 pKa = 3.6LLKK1136 pKa = 11.4GNGGRR1141 pKa = 11.84DD1142 pKa = 3.51NILGGDD1148 pKa = 3.88GNDD1151 pKa = 4.09SIEE1154 pKa = 4.34GGAGNDD1160 pKa = 3.24RR1161 pKa = 11.84LLGEE1165 pKa = 4.89SGDD1168 pKa = 3.91DD1169 pKa = 3.8TILGNAGQDD1178 pKa = 3.38TLDD1181 pKa = 3.78GGEE1184 pKa = 4.91GKK1186 pKa = 10.29DD1187 pKa = 3.68SLNGGTDD1194 pKa = 2.93NDD1196 pKa = 5.24LIFGGKK1202 pKa = 10.34GEE1204 pKa = 4.46DD1205 pKa = 3.67SLVGEE1210 pKa = 5.13AGADD1214 pKa = 3.61SLTGGASNDD1223 pKa = 3.89SLDD1226 pKa = 3.93GGAGSDD1232 pKa = 3.4RR1233 pKa = 11.84LVGVDD1238 pKa = 3.91PSASYY1243 pKa = 10.77GAGEE1247 pKa = 4.65LDD1249 pKa = 3.76VLTGFSDD1256 pKa = 3.19SDD1258 pKa = 3.85TFVLGDD1264 pKa = 3.79ASRR1267 pKa = 11.84VFYY1270 pKa = 11.11SDD1272 pKa = 4.28GDD1274 pKa = 3.9PLTTGEE1280 pKa = 4.45SDD1282 pKa = 3.62YY1283 pKa = 12.0ALISDD1288 pKa = 4.97FDD1290 pKa = 4.16SSQDD1294 pKa = 3.23IIQLHH1299 pKa = 5.7GAIAQYY1305 pKa = 11.26DD1306 pKa = 4.0LDD1308 pKa = 4.48FFTTSSGSINAALIYY1323 pKa = 10.67DD1324 pKa = 4.38PGDD1327 pKa = 3.33AARR1330 pKa = 11.84GEE1332 pKa = 4.73LISILQNVPTDD1343 pKa = 4.12LNLGSPAFSFVV1354 pKa = 2.94
Molecular weight: 142.62 kDa
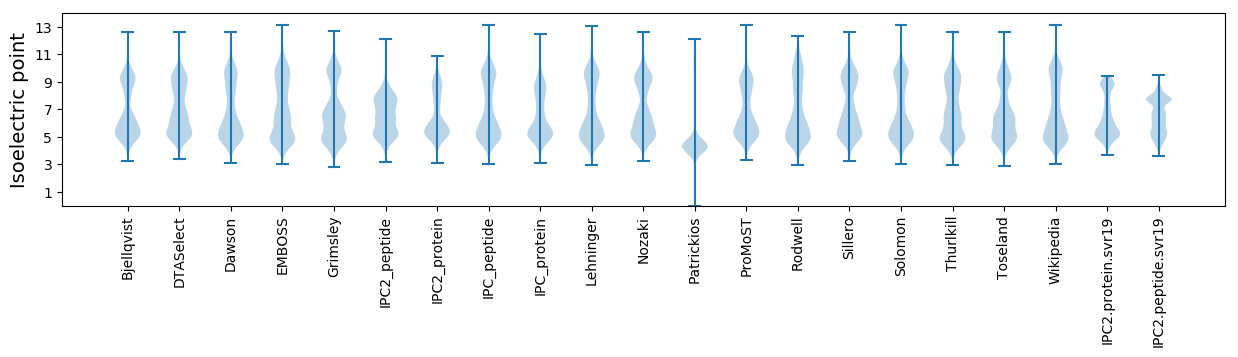
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9T3J3|K9T3J3_9CYAN Calcineurin-like phosphoesterase OS=Pleurocapsa sp. PCC 7327 OX=118163 GN=Ple7327_1620 PE=4 SV=1
MM1 pKa = 7.29TRR3 pKa = 11.84RR4 pKa = 11.84TLEE7 pKa = 3.73GTNRR11 pKa = 11.84KK12 pKa = 7.62QKK14 pKa = 8.83RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MKK24 pKa = 9.55TKK26 pKa = 10.27NGRR29 pKa = 11.84KK30 pKa = 8.82VLQARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.48GRR40 pKa = 11.84HH41 pKa = 4.07QLTVV45 pKa = 2.92
MM1 pKa = 7.29TRR3 pKa = 11.84RR4 pKa = 11.84TLEE7 pKa = 3.73GTNRR11 pKa = 11.84KK12 pKa = 7.62QKK14 pKa = 8.83RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MKK24 pKa = 9.55TKK26 pKa = 10.27NGRR29 pKa = 11.84KK30 pKa = 8.82VLQARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.48GRR40 pKa = 11.84HH41 pKa = 4.07QLTVV45 pKa = 2.92
Molecular weight: 5.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1342602 |
29 |
2921 |
315.5 |
35.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.574 ± 0.036 | 1.036 ± 0.014 |
4.819 ± 0.036 | 6.589 ± 0.04 |
3.957 ± 0.032 | 6.778 ± 0.041 |
1.691 ± 0.015 | 6.811 ± 0.031 |
4.768 ± 0.037 | 11.212 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.865 ± 0.017 | 3.83 ± 0.031 |
4.744 ± 0.025 | 4.975 ± 0.041 |
5.817 ± 0.028 | 6.274 ± 0.03 |
5.266 ± 0.027 | 6.477 ± 0.031 |
1.487 ± 0.019 | 3.029 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
