
Clostridium sp. CAG:1013
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
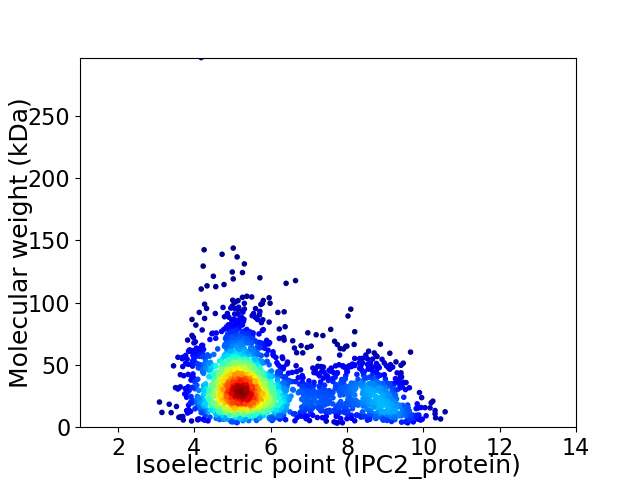
Virtual 2D-PAGE plot for 2437 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5A2M0|R5A2M0_9CLOT DNA polymerase III alpha subunit Gram-positive type OS=Clostridium sp. CAG:1013 OX=1262769 GN=BN452_01052 PE=4 SV=1
MM1 pKa = 7.33KK2 pKa = 10.28KK3 pKa = 9.88VLAMLLVFAMIASFAACGSSSNSSTVNEE31 pKa = 4.05EE32 pKa = 3.88SSAVSSDD39 pKa = 3.14KK40 pKa = 10.97VSSSTEE46 pKa = 4.08EE47 pKa = 3.84EE48 pKa = 4.4SSTGEE53 pKa = 4.13KK54 pKa = 10.52FSGSISIACSSQEE67 pKa = 3.64AKK69 pKa = 10.72ALQEE73 pKa = 3.95VLKK76 pKa = 10.72SYY78 pKa = 11.2NEE80 pKa = 3.91QEE82 pKa = 4.67DD83 pKa = 3.8VDD85 pKa = 4.62EE86 pKa = 4.34VTIDD90 pKa = 4.33PITVEE95 pKa = 4.09SVGDD99 pKa = 3.6FEE101 pKa = 6.39GLMTSWISSDD111 pKa = 4.07TLPDD115 pKa = 3.38MYY117 pKa = 10.45ISQVGSLQQSYY128 pKa = 10.09AANGYY133 pKa = 8.9LLPLTEE139 pKa = 4.52YY140 pKa = 11.07GFLDD144 pKa = 3.6RR145 pKa = 11.84LVAGDD150 pKa = 4.02TEE152 pKa = 4.83LIKK155 pKa = 10.9YY156 pKa = 10.46DD157 pKa = 3.7GDD159 pKa = 4.05DD160 pKa = 3.47YY161 pKa = 11.55AVPMNQSISVIYY173 pKa = 10.17VNNAALKK180 pKa = 9.29EE181 pKa = 3.99AGIEE185 pKa = 3.85IDD187 pKa = 3.33EE188 pKa = 4.45TNYY191 pKa = 10.3PNTWDD196 pKa = 3.79EE197 pKa = 4.57FVDD200 pKa = 4.49ILDD203 pKa = 3.65QLVAAGVQYY212 pKa = 10.4PFSLAGQDD220 pKa = 3.53ASAVTALPFQYY231 pKa = 9.71IYY233 pKa = 10.85QVVYY237 pKa = 10.01GANPNWYY244 pKa = 9.78ADD246 pKa = 3.58MLKK249 pKa = 10.74GEE251 pKa = 5.27NGAAWNDD258 pKa = 3.47EE259 pKa = 4.27LFLGMFDD266 pKa = 5.28KK267 pKa = 11.35YY268 pKa = 11.37GEE270 pKa = 4.01LLKK273 pKa = 11.01YY274 pKa = 10.63VSEE277 pKa = 4.56DD278 pKa = 3.29VLGVSKK284 pKa = 10.64DD285 pKa = 3.29EE286 pKa = 4.03RR287 pKa = 11.84LARR290 pKa = 11.84SVTGEE295 pKa = 3.79SPIYY299 pKa = 10.41VDD301 pKa = 3.96TSGAMPGIRR310 pKa = 11.84EE311 pKa = 4.66LDD313 pKa = 3.63PEE315 pKa = 5.75ADD317 pKa = 3.71VIMLPSCFVDD327 pKa = 5.34DD328 pKa = 5.22PADD331 pKa = 3.44QTIISSFDD339 pKa = 3.28SAVSITTGAEE349 pKa = 3.66KK350 pKa = 10.41KK351 pKa = 10.98GNVEE355 pKa = 4.06LCVDD359 pKa = 4.55FLDD362 pKa = 4.34YY363 pKa = 10.53LTTVEE368 pKa = 5.27GSTIFNNITGYY379 pKa = 10.7VPTIVDD385 pKa = 4.18CEE387 pKa = 4.69SEE389 pKa = 4.09VDD391 pKa = 3.55PAFDD395 pKa = 3.17IVLSILNKK403 pKa = 10.61GEE405 pKa = 4.37LPNSPMLSRR414 pKa = 11.84EE415 pKa = 4.27WIPGIKK421 pKa = 9.46EE422 pKa = 3.9VMKK425 pKa = 10.8NGCQNWLAGEE435 pKa = 4.35DD436 pKa = 3.93PKK438 pKa = 11.46SVADD442 pKa = 4.68TIQAEE447 pKa = 4.17QDD449 pKa = 3.4RR450 pKa = 11.84LEE452 pKa = 4.38EE453 pKa = 4.44ASPDD457 pKa = 3.27WVEE460 pKa = 5.76DD461 pKa = 3.71FLANYY466 pKa = 9.46NYY468 pKa = 10.59KK469 pKa = 10.65
MM1 pKa = 7.33KK2 pKa = 10.28KK3 pKa = 9.88VLAMLLVFAMIASFAACGSSSNSSTVNEE31 pKa = 4.05EE32 pKa = 3.88SSAVSSDD39 pKa = 3.14KK40 pKa = 10.97VSSSTEE46 pKa = 4.08EE47 pKa = 3.84EE48 pKa = 4.4SSTGEE53 pKa = 4.13KK54 pKa = 10.52FSGSISIACSSQEE67 pKa = 3.64AKK69 pKa = 10.72ALQEE73 pKa = 3.95VLKK76 pKa = 10.72SYY78 pKa = 11.2NEE80 pKa = 3.91QEE82 pKa = 4.67DD83 pKa = 3.8VDD85 pKa = 4.62EE86 pKa = 4.34VTIDD90 pKa = 4.33PITVEE95 pKa = 4.09SVGDD99 pKa = 3.6FEE101 pKa = 6.39GLMTSWISSDD111 pKa = 4.07TLPDD115 pKa = 3.38MYY117 pKa = 10.45ISQVGSLQQSYY128 pKa = 10.09AANGYY133 pKa = 8.9LLPLTEE139 pKa = 4.52YY140 pKa = 11.07GFLDD144 pKa = 3.6RR145 pKa = 11.84LVAGDD150 pKa = 4.02TEE152 pKa = 4.83LIKK155 pKa = 10.9YY156 pKa = 10.46DD157 pKa = 3.7GDD159 pKa = 4.05DD160 pKa = 3.47YY161 pKa = 11.55AVPMNQSISVIYY173 pKa = 10.17VNNAALKK180 pKa = 9.29EE181 pKa = 3.99AGIEE185 pKa = 3.85IDD187 pKa = 3.33EE188 pKa = 4.45TNYY191 pKa = 10.3PNTWDD196 pKa = 3.79EE197 pKa = 4.57FVDD200 pKa = 4.49ILDD203 pKa = 3.65QLVAAGVQYY212 pKa = 10.4PFSLAGQDD220 pKa = 3.53ASAVTALPFQYY231 pKa = 9.71IYY233 pKa = 10.85QVVYY237 pKa = 10.01GANPNWYY244 pKa = 9.78ADD246 pKa = 3.58MLKK249 pKa = 10.74GEE251 pKa = 5.27NGAAWNDD258 pKa = 3.47EE259 pKa = 4.27LFLGMFDD266 pKa = 5.28KK267 pKa = 11.35YY268 pKa = 11.37GEE270 pKa = 4.01LLKK273 pKa = 11.01YY274 pKa = 10.63VSEE277 pKa = 4.56DD278 pKa = 3.29VLGVSKK284 pKa = 10.64DD285 pKa = 3.29EE286 pKa = 4.03RR287 pKa = 11.84LARR290 pKa = 11.84SVTGEE295 pKa = 3.79SPIYY299 pKa = 10.41VDD301 pKa = 3.96TSGAMPGIRR310 pKa = 11.84EE311 pKa = 4.66LDD313 pKa = 3.63PEE315 pKa = 5.75ADD317 pKa = 3.71VIMLPSCFVDD327 pKa = 5.34DD328 pKa = 5.22PADD331 pKa = 3.44QTIISSFDD339 pKa = 3.28SAVSITTGAEE349 pKa = 3.66KK350 pKa = 10.41KK351 pKa = 10.98GNVEE355 pKa = 4.06LCVDD359 pKa = 4.55FLDD362 pKa = 4.34YY363 pKa = 10.53LTTVEE368 pKa = 5.27GSTIFNNITGYY379 pKa = 10.7VPTIVDD385 pKa = 4.18CEE387 pKa = 4.69SEE389 pKa = 4.09VDD391 pKa = 3.55PAFDD395 pKa = 3.17IVLSILNKK403 pKa = 10.61GEE405 pKa = 4.37LPNSPMLSRR414 pKa = 11.84EE415 pKa = 4.27WIPGIKK421 pKa = 9.46EE422 pKa = 3.9VMKK425 pKa = 10.8NGCQNWLAGEE435 pKa = 4.35DD436 pKa = 3.93PKK438 pKa = 11.46SVADD442 pKa = 4.68TIQAEE447 pKa = 4.17QDD449 pKa = 3.4RR450 pKa = 11.84LEE452 pKa = 4.38EE453 pKa = 4.44ASPDD457 pKa = 3.27WVEE460 pKa = 5.76DD461 pKa = 3.71FLANYY466 pKa = 9.46NYY468 pKa = 10.59KK469 pKa = 10.65
Molecular weight: 51.16 kDa
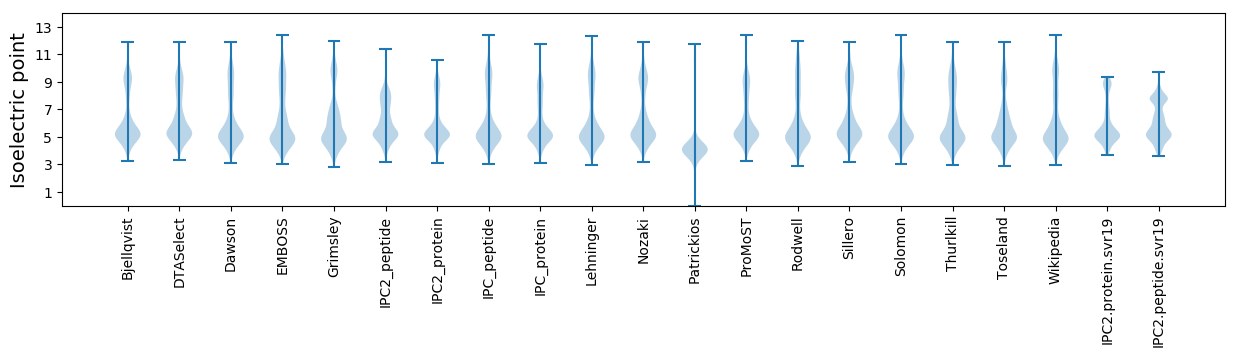
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5A769|R5A769_9CLOT N-acetylmuramoyl-L-alanine amidase OS=Clostridium sp. CAG:1013 OX=1262769 GN=BN452_01609 PE=4 SV=1
MM1 pKa = 7.19IQSKK5 pKa = 10.18LGRR8 pKa = 11.84ILGVVLLLAGLMVLSACVGSYY29 pKa = 10.11PLSPEE34 pKa = 4.52DD35 pKa = 2.63IWRR38 pKa = 11.84ILLGQGGDD46 pKa = 3.32TMEE49 pKa = 5.66ARR51 pKa = 11.84VFWQLRR57 pKa = 11.84LPRR60 pKa = 11.84VLMGVAAGAVMSLSGSVYY78 pKa = 7.49QTIFRR83 pKa = 11.84NPLASPDD90 pKa = 3.73LTGVASGASLGAAGAIVLGTGGAMQLMLGAFMAGMLSVVLVLFLVRR136 pKa = 11.84FSRR139 pKa = 11.84FRR141 pKa = 11.84QMSSYY146 pKa = 9.74ILAGIVVSSVADD158 pKa = 3.67AGLMCLKK165 pKa = 10.67VMADD169 pKa = 3.69PEE171 pKa = 4.29RR172 pKa = 11.84EE173 pKa = 3.71LAAIEE178 pKa = 4.35FWTMGSLGAITAGKK192 pKa = 9.95LFPNLLVMAVPFVLLLLFHH211 pKa = 6.83RR212 pKa = 11.84QAVMLSLGDD221 pKa = 3.58EE222 pKa = 4.56QARR225 pKa = 11.84SMGLNPSLWRR235 pKa = 11.84GIFLVLSTLMIAGLVSVTGVISFVGLIGPHH265 pKa = 4.84IAFWIRR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84RR274 pKa = 11.84DD275 pKa = 2.72RR276 pKa = 11.84GYY278 pKa = 11.07FLLSSALGAVLVLTADD294 pKa = 3.44MLARR298 pKa = 11.84SLWKK302 pKa = 10.42GAEE305 pKa = 4.12LPLSIFTVFLAVPILILLLRR325 pKa = 11.84GRR327 pKa = 11.84KK328 pKa = 9.24GDD330 pKa = 3.62FHH332 pKa = 6.3GTAAA336 pKa = 4.43
MM1 pKa = 7.19IQSKK5 pKa = 10.18LGRR8 pKa = 11.84ILGVVLLLAGLMVLSACVGSYY29 pKa = 10.11PLSPEE34 pKa = 4.52DD35 pKa = 2.63IWRR38 pKa = 11.84ILLGQGGDD46 pKa = 3.32TMEE49 pKa = 5.66ARR51 pKa = 11.84VFWQLRR57 pKa = 11.84LPRR60 pKa = 11.84VLMGVAAGAVMSLSGSVYY78 pKa = 7.49QTIFRR83 pKa = 11.84NPLASPDD90 pKa = 3.73LTGVASGASLGAAGAIVLGTGGAMQLMLGAFMAGMLSVVLVLFLVRR136 pKa = 11.84FSRR139 pKa = 11.84FRR141 pKa = 11.84QMSSYY146 pKa = 9.74ILAGIVVSSVADD158 pKa = 3.67AGLMCLKK165 pKa = 10.67VMADD169 pKa = 3.69PEE171 pKa = 4.29RR172 pKa = 11.84EE173 pKa = 3.71LAAIEE178 pKa = 4.35FWTMGSLGAITAGKK192 pKa = 9.95LFPNLLVMAVPFVLLLLFHH211 pKa = 6.83RR212 pKa = 11.84QAVMLSLGDD221 pKa = 3.58EE222 pKa = 4.56QARR225 pKa = 11.84SMGLNPSLWRR235 pKa = 11.84GIFLVLSTLMIAGLVSVTGVISFVGLIGPHH265 pKa = 4.84IAFWIRR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84RR274 pKa = 11.84DD275 pKa = 2.72RR276 pKa = 11.84GYY278 pKa = 11.07FLLSSALGAVLVLTADD294 pKa = 3.44MLARR298 pKa = 11.84SLWKK302 pKa = 10.42GAEE305 pKa = 4.12LPLSIFTVFLAVPILILLLRR325 pKa = 11.84GRR327 pKa = 11.84KK328 pKa = 9.24GDD330 pKa = 3.62FHH332 pKa = 6.3GTAAA336 pKa = 4.43
Molecular weight: 35.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
752331 |
29 |
2703 |
308.7 |
34.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.141 ± 0.05 | 1.637 ± 0.023 |
5.382 ± 0.041 | 7.303 ± 0.057 |
4.225 ± 0.038 | 7.872 ± 0.05 |
1.894 ± 0.027 | 5.506 ± 0.046 |
5.14 ± 0.045 | 10.107 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.746 ± 0.025 | 3.404 ± 0.031 |
4.289 ± 0.037 | 3.92 ± 0.032 |
5.03 ± 0.049 | 5.809 ± 0.048 |
5.423 ± 0.043 | 7.446 ± 0.043 |
1.137 ± 0.021 | 3.579 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
