
Eidolon helvum papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Psipapillomavirus; Psipapillomavirus 2
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
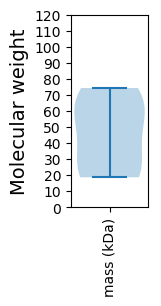
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1P8YVV0|A0A1P8YVV0_9PAPI Replication protein E1 OS=Eidolon helvum papillomavirus 2 OX=1335476 GN=E1 PE=3 SV=1
MM1 pKa = 7.57PAAKK5 pKa = 9.57RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84VRR10 pKa = 11.84RR11 pKa = 11.84EE12 pKa = 3.3TVYY15 pKa = 10.73RR16 pKa = 11.84RR17 pKa = 11.84VPRR20 pKa = 11.84DD21 pKa = 3.14TRR23 pKa = 11.84SRR25 pKa = 11.84RR26 pKa = 11.84VIRR29 pKa = 11.84DD30 pKa = 3.28SVGNIYY36 pKa = 10.37RR37 pKa = 11.84NCKK40 pKa = 8.32QWGTCPPDD48 pKa = 3.56VINKK52 pKa = 9.1VEE54 pKa = 4.23QKK56 pKa = 9.6TPADD60 pKa = 4.1NILKK64 pKa = 9.41YY65 pKa = 10.64GSSAVFFGGLGIGTGRR81 pKa = 11.84GSGGAGGYY89 pKa = 9.56IPLGSGGGVRR99 pKa = 11.84VGSWPWVPRR108 pKa = 11.84YY109 pKa = 7.87TGPTVEE115 pKa = 4.99SIGPATIPDD124 pKa = 3.71VTGSGWPWLPTYY136 pKa = 8.6PTEE139 pKa = 4.21AAGAGEE145 pKa = 4.4TIEE148 pKa = 5.55LEE150 pKa = 4.49TILPTNTTVDD160 pKa = 3.84EE161 pKa = 4.73TPVVAPDD168 pKa = 3.4APAVVDD174 pKa = 4.05EE175 pKa = 4.57LTNYY179 pKa = 7.45PQQPTVIDD187 pKa = 3.59STQPTTGGGRR197 pKa = 11.84VIDD200 pKa = 4.05VVAEE204 pKa = 3.88VHH206 pKa = 6.56HH207 pKa = 6.61PVDD210 pKa = 4.59FLPEE214 pKa = 3.93HH215 pKa = 5.78TLEE218 pKa = 4.63SGPSVGSEE226 pKa = 3.67PAVLDD231 pKa = 3.63VGEE234 pKa = 5.18EE235 pKa = 3.71IPMFPRR241 pKa = 11.84SRR243 pKa = 11.84IHH245 pKa = 7.15DD246 pKa = 3.84HH247 pKa = 6.71LEE249 pKa = 3.57PSILHH254 pKa = 5.67GTTSRR259 pKa = 11.84GTSADD264 pKa = 3.09VGGRR268 pKa = 11.84FSIIDD273 pKa = 3.43SSVAEE278 pKa = 4.12TFIGEE283 pKa = 4.35EE284 pKa = 4.64PYY286 pKa = 9.4STTPVRR292 pKa = 11.84YY293 pKa = 9.81DD294 pKa = 3.43PEE296 pKa = 4.2EE297 pKa = 3.95PLQFRR302 pKa = 11.84TSTPEE307 pKa = 3.68TSRR310 pKa = 11.84GPVARR315 pKa = 11.84LKK317 pKa = 10.75ALYY320 pKa = 10.24NRR322 pKa = 11.84FVQQIPVEE330 pKa = 4.41DD331 pKa = 4.15PLFVEE336 pKa = 4.7APGGFVEE343 pKa = 4.67YY344 pKa = 10.44DD345 pKa = 3.21NPAYY349 pKa = 10.64DD350 pKa = 3.99PDD352 pKa = 3.57ATIEE356 pKa = 4.19FNTEE360 pKa = 3.52DD361 pKa = 3.67ALEE364 pKa = 4.27ATVAPDD370 pKa = 3.33TRR372 pKa = 11.84LSDD375 pKa = 3.55LRR377 pKa = 11.84VLHH380 pKa = 6.85RR381 pKa = 11.84PLLSEE386 pKa = 4.1TPTGHH391 pKa = 5.84VRR393 pKa = 11.84VSRR396 pKa = 11.84LGNFATMRR404 pKa = 11.84LRR406 pKa = 11.84SGAIPGPRR414 pKa = 11.84AHH416 pKa = 6.88VYY418 pKa = 10.54HH419 pKa = 7.32DD420 pKa = 4.17LSSIGEE426 pKa = 4.33SFEE429 pKa = 3.98LTPFGSGRR437 pKa = 11.84VSASTPVVLDD447 pKa = 3.55TPVDD451 pKa = 3.97VVVDD455 pKa = 3.84TPVDD459 pKa = 3.6VLTEE463 pKa = 4.08DD464 pKa = 3.46TTGLEE469 pKa = 4.72DD470 pKa = 3.86GLEE473 pKa = 4.14DD474 pKa = 5.41VPLLDD479 pKa = 4.48HH480 pKa = 7.32GSGEE484 pKa = 4.23VQRR487 pKa = 11.84VGVGRR492 pKa = 11.84GGSTNRR498 pKa = 11.84SFVSMEE504 pKa = 3.91LPSTGRR510 pKa = 11.84SDD512 pKa = 4.34GFGTTISDD520 pKa = 3.56IDD522 pKa = 3.54SDD524 pKa = 4.78AYY526 pKa = 10.31VHH528 pKa = 6.35YY529 pKa = 8.42PTDD532 pKa = 3.32STEE535 pKa = 3.84PVAPSFGPGIPVEE548 pKa = 4.07PAGPSIDD555 pKa = 3.79VGDD558 pKa = 3.72VGIDD562 pKa = 3.66YY563 pKa = 10.9FLDD566 pKa = 4.57PYY568 pKa = 10.99LFRR571 pKa = 11.84KK572 pKa = 8.7RR573 pKa = 11.84RR574 pKa = 11.84KK575 pKa = 8.51RR576 pKa = 11.84KK577 pKa = 9.63RR578 pKa = 11.84FFSFADD584 pKa = 3.46DD585 pKa = 4.21HH586 pKa = 7.01VDD588 pKa = 3.18SS589 pKa = 4.89
MM1 pKa = 7.57PAAKK5 pKa = 9.57RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84VRR10 pKa = 11.84RR11 pKa = 11.84EE12 pKa = 3.3TVYY15 pKa = 10.73RR16 pKa = 11.84RR17 pKa = 11.84VPRR20 pKa = 11.84DD21 pKa = 3.14TRR23 pKa = 11.84SRR25 pKa = 11.84RR26 pKa = 11.84VIRR29 pKa = 11.84DD30 pKa = 3.28SVGNIYY36 pKa = 10.37RR37 pKa = 11.84NCKK40 pKa = 8.32QWGTCPPDD48 pKa = 3.56VINKK52 pKa = 9.1VEE54 pKa = 4.23QKK56 pKa = 9.6TPADD60 pKa = 4.1NILKK64 pKa = 9.41YY65 pKa = 10.64GSSAVFFGGLGIGTGRR81 pKa = 11.84GSGGAGGYY89 pKa = 9.56IPLGSGGGVRR99 pKa = 11.84VGSWPWVPRR108 pKa = 11.84YY109 pKa = 7.87TGPTVEE115 pKa = 4.99SIGPATIPDD124 pKa = 3.71VTGSGWPWLPTYY136 pKa = 8.6PTEE139 pKa = 4.21AAGAGEE145 pKa = 4.4TIEE148 pKa = 5.55LEE150 pKa = 4.49TILPTNTTVDD160 pKa = 3.84EE161 pKa = 4.73TPVVAPDD168 pKa = 3.4APAVVDD174 pKa = 4.05EE175 pKa = 4.57LTNYY179 pKa = 7.45PQQPTVIDD187 pKa = 3.59STQPTTGGGRR197 pKa = 11.84VIDD200 pKa = 4.05VVAEE204 pKa = 3.88VHH206 pKa = 6.56HH207 pKa = 6.61PVDD210 pKa = 4.59FLPEE214 pKa = 3.93HH215 pKa = 5.78TLEE218 pKa = 4.63SGPSVGSEE226 pKa = 3.67PAVLDD231 pKa = 3.63VGEE234 pKa = 5.18EE235 pKa = 3.71IPMFPRR241 pKa = 11.84SRR243 pKa = 11.84IHH245 pKa = 7.15DD246 pKa = 3.84HH247 pKa = 6.71LEE249 pKa = 3.57PSILHH254 pKa = 5.67GTTSRR259 pKa = 11.84GTSADD264 pKa = 3.09VGGRR268 pKa = 11.84FSIIDD273 pKa = 3.43SSVAEE278 pKa = 4.12TFIGEE283 pKa = 4.35EE284 pKa = 4.64PYY286 pKa = 9.4STTPVRR292 pKa = 11.84YY293 pKa = 9.81DD294 pKa = 3.43PEE296 pKa = 4.2EE297 pKa = 3.95PLQFRR302 pKa = 11.84TSTPEE307 pKa = 3.68TSRR310 pKa = 11.84GPVARR315 pKa = 11.84LKK317 pKa = 10.75ALYY320 pKa = 10.24NRR322 pKa = 11.84FVQQIPVEE330 pKa = 4.41DD331 pKa = 4.15PLFVEE336 pKa = 4.7APGGFVEE343 pKa = 4.67YY344 pKa = 10.44DD345 pKa = 3.21NPAYY349 pKa = 10.64DD350 pKa = 3.99PDD352 pKa = 3.57ATIEE356 pKa = 4.19FNTEE360 pKa = 3.52DD361 pKa = 3.67ALEE364 pKa = 4.27ATVAPDD370 pKa = 3.33TRR372 pKa = 11.84LSDD375 pKa = 3.55LRR377 pKa = 11.84VLHH380 pKa = 6.85RR381 pKa = 11.84PLLSEE386 pKa = 4.1TPTGHH391 pKa = 5.84VRR393 pKa = 11.84VSRR396 pKa = 11.84LGNFATMRR404 pKa = 11.84LRR406 pKa = 11.84SGAIPGPRR414 pKa = 11.84AHH416 pKa = 6.88VYY418 pKa = 10.54HH419 pKa = 7.32DD420 pKa = 4.17LSSIGEE426 pKa = 4.33SFEE429 pKa = 3.98LTPFGSGRR437 pKa = 11.84VSASTPVVLDD447 pKa = 3.55TPVDD451 pKa = 3.97VVVDD455 pKa = 3.84TPVDD459 pKa = 3.6VLTEE463 pKa = 4.08DD464 pKa = 3.46TTGLEE469 pKa = 4.72DD470 pKa = 3.86GLEE473 pKa = 4.14DD474 pKa = 5.41VPLLDD479 pKa = 4.48HH480 pKa = 7.32GSGEE484 pKa = 4.23VQRR487 pKa = 11.84VGVGRR492 pKa = 11.84GGSTNRR498 pKa = 11.84SFVSMEE504 pKa = 3.91LPSTGRR510 pKa = 11.84SDD512 pKa = 4.34GFGTTISDD520 pKa = 3.56IDD522 pKa = 3.54SDD524 pKa = 4.78AYY526 pKa = 10.31VHH528 pKa = 6.35YY529 pKa = 8.42PTDD532 pKa = 3.32STEE535 pKa = 3.84PVAPSFGPGIPVEE548 pKa = 4.07PAGPSIDD555 pKa = 3.79VGDD558 pKa = 3.72VGIDD562 pKa = 3.66YY563 pKa = 10.9FLDD566 pKa = 4.57PYY568 pKa = 10.99LFRR571 pKa = 11.84KK572 pKa = 8.7RR573 pKa = 11.84RR574 pKa = 11.84KK575 pKa = 8.51RR576 pKa = 11.84KK577 pKa = 9.63RR578 pKa = 11.84FFSFADD584 pKa = 3.46DD585 pKa = 4.21HH586 pKa = 7.01VDD588 pKa = 3.18SS589 pKa = 4.89
Molecular weight: 63.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8YVV8|A0A1P8YVV8_9PAPI Protein E6 OS=Eidolon helvum papillomavirus 2 OX=1335476 GN=E6 PE=3 SV=1
MM1 pKa = 7.38LTTFFIFRR9 pKa = 11.84EE10 pKa = 4.45SKK12 pKa = 9.68TDD14 pKa = 3.4TGCGRR19 pKa = 11.84SIKK22 pKa = 10.64GPLRR26 pKa = 11.84LPFAPMAHH34 pKa = 7.07PEE36 pKa = 4.41SITDD40 pKa = 3.98LARR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84GTTVRR50 pKa = 11.84DD51 pKa = 3.26ITIPCGFCNEE61 pKa = 4.53PLNDD65 pKa = 3.42QDD67 pKa = 5.56KK68 pKa = 10.71YY69 pKa = 11.49AFDD72 pKa = 3.95VRR74 pKa = 11.84NHH76 pKa = 6.43RR77 pKa = 11.84LLNRR81 pKa = 11.84RR82 pKa = 11.84GVYY85 pKa = 9.97FGCCTPCVRR94 pKa = 11.84ALARR98 pKa = 11.84KK99 pKa = 9.89DD100 pKa = 3.45RR101 pKa = 11.84DD102 pKa = 3.74TGNLAPFTVAEE113 pKa = 4.17AEE115 pKa = 4.26AYY117 pKa = 9.09VGKK120 pKa = 10.32NIGQILVLCSCCLKK134 pKa = 10.76RR135 pKa = 11.84LDD137 pKa = 5.2LIEE140 pKa = 4.63KK141 pKa = 9.91LIHH144 pKa = 5.49QRR146 pKa = 11.84AGFYY150 pKa = 10.75LCMVRR155 pKa = 11.84DD156 pKa = 3.9GEE158 pKa = 4.25FRR160 pKa = 11.84GICRR164 pKa = 11.84LCMVLEE170 pKa = 4.39GQEE173 pKa = 3.84
MM1 pKa = 7.38LTTFFIFRR9 pKa = 11.84EE10 pKa = 4.45SKK12 pKa = 9.68TDD14 pKa = 3.4TGCGRR19 pKa = 11.84SIKK22 pKa = 10.64GPLRR26 pKa = 11.84LPFAPMAHH34 pKa = 7.07PEE36 pKa = 4.41SITDD40 pKa = 3.98LARR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84GTTVRR50 pKa = 11.84DD51 pKa = 3.26ITIPCGFCNEE61 pKa = 4.53PLNDD65 pKa = 3.42QDD67 pKa = 5.56KK68 pKa = 10.71YY69 pKa = 11.49AFDD72 pKa = 3.95VRR74 pKa = 11.84NHH76 pKa = 6.43RR77 pKa = 11.84LLNRR81 pKa = 11.84RR82 pKa = 11.84GVYY85 pKa = 9.97FGCCTPCVRR94 pKa = 11.84ALARR98 pKa = 11.84KK99 pKa = 9.89DD100 pKa = 3.45RR101 pKa = 11.84DD102 pKa = 3.74TGNLAPFTVAEE113 pKa = 4.17AEE115 pKa = 4.26AYY117 pKa = 9.09VGKK120 pKa = 10.32NIGQILVLCSCCLKK134 pKa = 10.76RR135 pKa = 11.84LDD137 pKa = 5.2LIEE140 pKa = 4.63KK141 pKa = 9.91LIHH144 pKa = 5.49QRR146 pKa = 11.84AGFYY150 pKa = 10.75LCMVRR155 pKa = 11.84DD156 pKa = 3.9GEE158 pKa = 4.25FRR160 pKa = 11.84GICRR164 pKa = 11.84LCMVLEE170 pKa = 4.39GQEE173 pKa = 3.84
Molecular weight: 19.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2439 |
166 |
663 |
406.5 |
45.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.56 ± 0.981 | 2.132 ± 0.633 |
6.191 ± 0.476 | 6.642 ± 0.402 |
3.526 ± 0.225 | 7.708 ± 0.916 |
2.009 ± 0.165 | 4.059 ± 0.296 |
3.649 ± 0.745 | 7.831 ± 0.591 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.091 ± 0.517 | 3.198 ± 0.732 |
6.765 ± 1.139 | 4.223 ± 0.896 |
7.339 ± 0.926 | 6.56 ± 0.573 |
7.503 ± 0.516 | 7.298 ± 0.934 |
1.312 ± 0.262 | 3.403 ± 0.335 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
