
Sanxia water strider virus 19
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.38
Get precalculated fractions of proteins
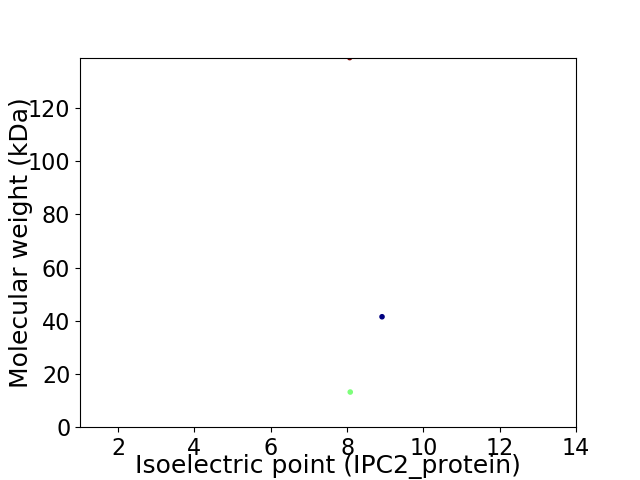
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
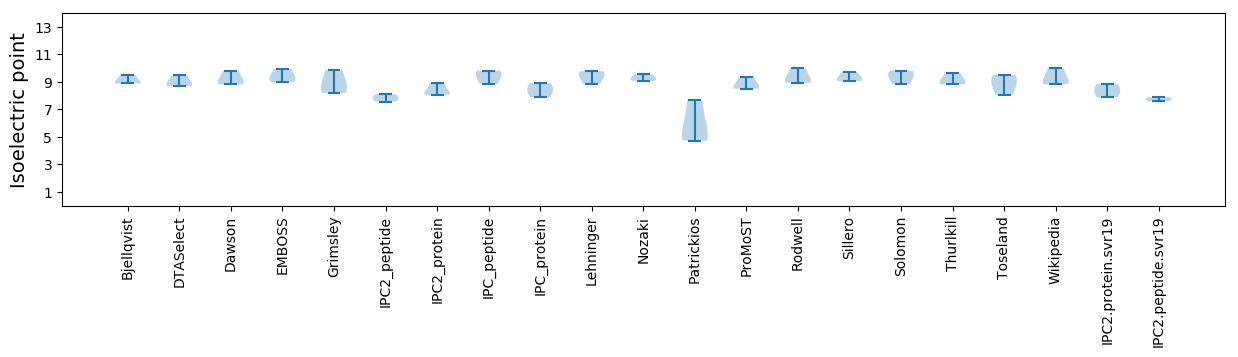
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHR7|A0A1L3KHR7_9VIRU Uncharacterized protein OS=Sanxia water strider virus 19 OX=1923403 PE=4 SV=1
MM1 pKa = 7.51APIPTHH7 pKa = 6.7LLTPFSDD14 pKa = 2.92NHH16 pKa = 6.41LAFQTSIMDD25 pKa = 3.95ASNPVLDD32 pKa = 4.29SSRR35 pKa = 11.84MTIRR39 pKa = 11.84SMMEE43 pKa = 2.99KK44 pKa = 9.42HH45 pKa = 6.7ARR47 pKa = 11.84RR48 pKa = 11.84QRR50 pKa = 11.84QEE52 pKa = 3.36LSYY55 pKa = 10.35VARR58 pKa = 11.84LAEE61 pKa = 4.31EE62 pKa = 4.23YY63 pKa = 10.37PRR65 pKa = 11.84VRR67 pKa = 11.84FGKK70 pKa = 10.42GVAKK74 pKa = 10.33EE75 pKa = 3.88MSRR78 pKa = 11.84EE79 pKa = 4.03DD80 pKa = 4.62KK81 pKa = 9.89ATAWAQIEE89 pKa = 4.54RR90 pKa = 11.84KK91 pKa = 7.59TPQLDD96 pKa = 3.59TSPSLDD102 pKa = 3.57LDD104 pKa = 3.81SFLDD108 pKa = 3.73ARR110 pKa = 11.84PPCPVSQVSYY120 pKa = 10.52HH121 pKa = 5.94SVPLHH126 pKa = 6.57PEE128 pKa = 3.76NTCRR132 pKa = 11.84SGPHH136 pKa = 5.04VSATRR141 pKa = 11.84MFGQIYY147 pKa = 9.83KK148 pKa = 10.4DD149 pKa = 3.14KK150 pKa = 11.36LMTDD154 pKa = 3.44RR155 pKa = 11.84LLALEE160 pKa = 4.29PEE162 pKa = 4.79FVKK165 pKa = 10.36EE166 pKa = 3.82HH167 pKa = 6.93YY168 pKa = 9.98YY169 pKa = 10.93DD170 pKa = 3.42QVYY173 pKa = 11.15SDD175 pKa = 3.75GTAQGLLNRR184 pKa = 11.84MRR186 pKa = 11.84KK187 pKa = 9.02QMSNKK192 pKa = 7.54VTPLSQLEE200 pKa = 4.38KK201 pKa = 9.98VAKK204 pKa = 9.67TNLSTKK210 pKa = 10.37RR211 pKa = 11.84MSTLIGEE218 pKa = 4.44MFPRR222 pKa = 11.84KK223 pKa = 8.6KK224 pKa = 8.99TLPDD228 pKa = 2.89WHH230 pKa = 7.39RR231 pKa = 11.84DD232 pKa = 2.95LDD234 pKa = 3.99LQLEE238 pKa = 4.44EE239 pKa = 4.58VVQSAKK245 pKa = 10.75ASAGFPYY252 pKa = 9.98CRR254 pKa = 11.84PKK256 pKa = 11.3GEE258 pKa = 4.21VLDD261 pKa = 3.98QVVCAGIPLLCDD273 pKa = 4.9AIRR276 pKa = 11.84EE277 pKa = 4.28GKK279 pKa = 9.82LAEE282 pKa = 4.08FRR284 pKa = 11.84AANPEE289 pKa = 3.91FFLCEE294 pKa = 4.08VKK296 pKa = 10.69NKK298 pKa = 9.43RR299 pKa = 11.84DD300 pKa = 3.38RR301 pKa = 11.84YY302 pKa = 7.66EE303 pKa = 3.79TAKK306 pKa = 9.15ITEE309 pKa = 3.96KK310 pKa = 9.91TRR312 pKa = 11.84PYY314 pKa = 10.68VAFPAHH320 pKa = 5.85FQLLFSCLSQPFTKK334 pKa = 10.48DD335 pKa = 2.7LLLFHH340 pKa = 6.84EE341 pKa = 5.23HH342 pKa = 6.6PKK344 pKa = 10.61SVNAYY349 pKa = 8.77GFTMAHH355 pKa = 6.37GGSSKK360 pKa = 9.92IRR362 pKa = 11.84YY363 pKa = 6.49RR364 pKa = 11.84TAEE367 pKa = 3.86LRR369 pKa = 11.84EE370 pKa = 4.26LGSKK374 pKa = 10.29GGAPICRR381 pKa = 11.84HH382 pKa = 5.96YY383 pKa = 11.41GDD385 pKa = 5.16DD386 pKa = 3.23ADD388 pKa = 3.55IYY390 pKa = 10.24VRR392 pKa = 11.84RR393 pKa = 11.84GGKK396 pKa = 8.88LLRR399 pKa = 11.84LCPDD403 pKa = 4.16FSQMDD408 pKa = 3.69GSVTHH413 pKa = 5.4EE414 pKa = 4.24TVRR417 pKa = 11.84LTVDD421 pKa = 4.06WILASYY427 pKa = 9.26EE428 pKa = 4.29EE429 pKa = 4.21QWGPSDD435 pKa = 3.66FWRR438 pKa = 11.84TIGDD442 pKa = 3.72LWVEE446 pKa = 4.32MATSPLFVVHH456 pKa = 7.08GKK458 pKa = 9.67DD459 pKa = 3.04VWEE462 pKa = 4.81KK463 pKa = 9.12RR464 pKa = 11.84TQDD467 pKa = 3.32GLMTGVVGTTLFDD480 pKa = 3.66TVHH483 pKa = 6.69AALAYY488 pKa = 7.21WVWVEE493 pKa = 3.78RR494 pKa = 11.84VHH496 pKa = 6.29YY497 pKa = 9.28QRR499 pKa = 11.84QYY501 pKa = 11.88ALLEE505 pKa = 3.88EE506 pKa = 4.6APAIKK511 pKa = 10.18LFKK514 pKa = 10.98EE515 pKa = 3.7MGLVVKK521 pKa = 10.3PGTWGFEE528 pKa = 4.26TVNEE532 pKa = 4.13NPVQGTLWTTQRR544 pKa = 11.84FLGVQYY550 pKa = 10.59KK551 pKa = 9.02WEE553 pKa = 4.17TGPRR557 pKa = 11.84EE558 pKa = 4.42VEE560 pKa = 4.33MVPHH564 pKa = 6.92LAYY567 pKa = 10.18EE568 pKa = 4.32DD569 pKa = 3.78WLSLLLCPRR578 pKa = 11.84GDD580 pKa = 3.34EE581 pKa = 4.41DD582 pKa = 3.61MRR584 pKa = 11.84SSRR587 pKa = 11.84TSEE590 pKa = 3.76LTRR593 pKa = 11.84EE594 pKa = 3.95RR595 pKa = 11.84VRR597 pKa = 11.84FDD599 pKa = 3.34RR600 pKa = 11.84LRR602 pKa = 11.84GMIVTGAFSTPEE614 pKa = 3.72ALAIMYY620 pKa = 8.39EE621 pKa = 4.55LIDD624 pKa = 3.9EE625 pKa = 4.57VNPVAIIMDD634 pKa = 3.86VQSGDD639 pKa = 3.74GKK641 pKa = 11.19GVTPDD646 pKa = 4.74AIALLNEE653 pKa = 4.27DD654 pKa = 4.2FQWPGSEE661 pKa = 5.62GVPDD665 pKa = 4.31WKK667 pKa = 10.46WCMNLYY673 pKa = 10.44LSPEE677 pKa = 3.98NQYY680 pKa = 11.39EE681 pKa = 3.97DD682 pKa = 4.4AQWKK686 pKa = 9.45LVIPEE691 pKa = 3.95AHH693 pKa = 6.74AALSDD698 pKa = 3.43FKK700 pKa = 11.41VRR702 pKa = 11.84YY703 pKa = 9.79RR704 pKa = 11.84KK705 pKa = 9.56DD706 pKa = 3.35LKK708 pKa = 11.04KK709 pKa = 10.57LATVQKK715 pKa = 10.11DD716 pKa = 3.81VEE718 pKa = 4.56GKK720 pKa = 9.48PSAAAMLPIPDD731 pKa = 5.16PAPKK735 pKa = 10.26AQIPSMEE742 pKa = 4.27IPTSGSRR749 pKa = 11.84VPKK752 pKa = 10.17TSEE755 pKa = 4.11AYY757 pKa = 10.49NKK759 pKa = 6.78TTKK762 pKa = 10.39PVDD765 pKa = 3.41VTLDD769 pKa = 3.63DD770 pKa = 3.25QGLAVEE776 pKa = 4.42EE777 pKa = 4.29RR778 pKa = 11.84TRR780 pKa = 11.84RR781 pKa = 11.84PTDD784 pKa = 3.12VEE786 pKa = 4.32QILSTFRR793 pKa = 11.84DD794 pKa = 3.72LPLSPKK800 pKa = 9.78AWEE803 pKa = 4.55EE804 pKa = 3.83IVAGATEE811 pKa = 4.12TQRR814 pKa = 11.84AGLEE818 pKa = 3.96ALQGGTKK825 pKa = 9.7VVQPVFQASTLSWKK839 pKa = 10.71LNWTVSKK846 pKa = 10.89VLSTAQKK853 pKa = 10.03AGLVVLPGGPKK864 pKa = 9.8RR865 pKa = 11.84KK866 pKa = 9.87SLITALPPSDD876 pKa = 4.1PSALEE881 pKa = 4.91PIDD884 pKa = 3.73HH885 pKa = 6.92AAAAVEE891 pKa = 3.99RR892 pKa = 11.84ALDD895 pKa = 3.63NSRR898 pKa = 11.84PEE900 pKa = 3.82PKK902 pKa = 9.32EE903 pKa = 3.32WSRR906 pKa = 11.84EE907 pKa = 3.86HH908 pKa = 6.21SQLGLSRR915 pKa = 11.84LKK917 pKa = 11.13GPLTQEE923 pKa = 3.84QAAKK927 pKa = 9.81VCKK930 pKa = 9.94EE931 pKa = 3.84FLAHH935 pKa = 7.78GYY937 pKa = 8.73RR938 pKa = 11.84CPMPKK943 pKa = 10.1CKK945 pKa = 10.02FGKK948 pKa = 10.51GNAAPLTFVEE958 pKa = 5.32VTLHH962 pKa = 6.98WITNDD967 pKa = 3.14PHH969 pKa = 7.38QDD971 pKa = 2.81KK972 pKa = 11.0CLLSKK977 pKa = 11.28CEE979 pKa = 4.22LHH981 pKa = 7.21DD982 pKa = 4.28GCPTGPPMQLRR993 pKa = 11.84PAKK996 pKa = 10.24EE997 pKa = 3.29ITLLKK1002 pKa = 10.16KK1003 pKa = 9.9QAEE1006 pKa = 4.31RR1007 pKa = 11.84LDD1009 pKa = 4.11GPQAEE1014 pKa = 4.69VLPPYY1019 pKa = 9.41PRR1021 pKa = 11.84PPLKK1025 pKa = 10.3FSSMDD1030 pKa = 3.35LEE1032 pKa = 4.77EE1033 pKa = 4.28IRR1035 pKa = 11.84SIQAEE1040 pKa = 4.58DD1041 pKa = 3.53PSMARR1046 pKa = 11.84SVFSSMCHH1054 pKa = 4.34EE1055 pKa = 4.18QGVAVVFTTKK1065 pKa = 9.83GTDD1068 pKa = 3.31HH1069 pKa = 7.07EE1070 pKa = 4.76KK1071 pKa = 10.98GAVNAPTYY1079 pKa = 11.39GEE1081 pKa = 4.16MHH1083 pKa = 6.14WTRR1086 pKa = 11.84IKK1088 pKa = 10.82KK1089 pKa = 9.65QGQPTPSFKK1098 pKa = 10.58RR1099 pKa = 11.84EE1100 pKa = 3.59PLAWNLYY1107 pKa = 9.47LQVLGKK1113 pKa = 10.17KK1114 pKa = 9.7RR1115 pKa = 11.84EE1116 pKa = 4.04LHH1118 pKa = 5.96NVLLTTALASLSDD1131 pKa = 3.89LGKK1134 pKa = 9.88PSLPPASWYY1143 pKa = 9.24EE1144 pKa = 3.8AAKK1147 pKa = 10.96VEE1149 pKa = 4.4LLPTASKK1156 pKa = 10.63AYY1158 pKa = 9.26STVKK1162 pKa = 10.08TPAPVLLNSKK1172 pKa = 9.65PSKK1175 pKa = 9.79TPLSQSISHH1184 pKa = 7.0AYY1186 pKa = 6.34EE1187 pKa = 4.05TKK1189 pKa = 10.08PQPSKK1194 pKa = 10.86PKK1196 pKa = 9.68TEE1198 pKa = 4.39SKK1200 pKa = 10.51PISKK1204 pKa = 9.06WGKK1207 pKa = 9.37KK1208 pKa = 9.48KK1209 pKa = 9.88PAAPGQAKK1217 pKa = 9.75KK1218 pKa = 10.72KK1219 pKa = 9.97KK1220 pKa = 9.02GPKK1223 pKa = 8.58KK1224 pKa = 9.65PSYY1227 pKa = 9.91GRR1229 pKa = 11.84SGATGSLPNRR1239 pKa = 11.84NSQQ1242 pKa = 3.15
MM1 pKa = 7.51APIPTHH7 pKa = 6.7LLTPFSDD14 pKa = 2.92NHH16 pKa = 6.41LAFQTSIMDD25 pKa = 3.95ASNPVLDD32 pKa = 4.29SSRR35 pKa = 11.84MTIRR39 pKa = 11.84SMMEE43 pKa = 2.99KK44 pKa = 9.42HH45 pKa = 6.7ARR47 pKa = 11.84RR48 pKa = 11.84QRR50 pKa = 11.84QEE52 pKa = 3.36LSYY55 pKa = 10.35VARR58 pKa = 11.84LAEE61 pKa = 4.31EE62 pKa = 4.23YY63 pKa = 10.37PRR65 pKa = 11.84VRR67 pKa = 11.84FGKK70 pKa = 10.42GVAKK74 pKa = 10.33EE75 pKa = 3.88MSRR78 pKa = 11.84EE79 pKa = 4.03DD80 pKa = 4.62KK81 pKa = 9.89ATAWAQIEE89 pKa = 4.54RR90 pKa = 11.84KK91 pKa = 7.59TPQLDD96 pKa = 3.59TSPSLDD102 pKa = 3.57LDD104 pKa = 3.81SFLDD108 pKa = 3.73ARR110 pKa = 11.84PPCPVSQVSYY120 pKa = 10.52HH121 pKa = 5.94SVPLHH126 pKa = 6.57PEE128 pKa = 3.76NTCRR132 pKa = 11.84SGPHH136 pKa = 5.04VSATRR141 pKa = 11.84MFGQIYY147 pKa = 9.83KK148 pKa = 10.4DD149 pKa = 3.14KK150 pKa = 11.36LMTDD154 pKa = 3.44RR155 pKa = 11.84LLALEE160 pKa = 4.29PEE162 pKa = 4.79FVKK165 pKa = 10.36EE166 pKa = 3.82HH167 pKa = 6.93YY168 pKa = 9.98YY169 pKa = 10.93DD170 pKa = 3.42QVYY173 pKa = 11.15SDD175 pKa = 3.75GTAQGLLNRR184 pKa = 11.84MRR186 pKa = 11.84KK187 pKa = 9.02QMSNKK192 pKa = 7.54VTPLSQLEE200 pKa = 4.38KK201 pKa = 9.98VAKK204 pKa = 9.67TNLSTKK210 pKa = 10.37RR211 pKa = 11.84MSTLIGEE218 pKa = 4.44MFPRR222 pKa = 11.84KK223 pKa = 8.6KK224 pKa = 8.99TLPDD228 pKa = 2.89WHH230 pKa = 7.39RR231 pKa = 11.84DD232 pKa = 2.95LDD234 pKa = 3.99LQLEE238 pKa = 4.44EE239 pKa = 4.58VVQSAKK245 pKa = 10.75ASAGFPYY252 pKa = 9.98CRR254 pKa = 11.84PKK256 pKa = 11.3GEE258 pKa = 4.21VLDD261 pKa = 3.98QVVCAGIPLLCDD273 pKa = 4.9AIRR276 pKa = 11.84EE277 pKa = 4.28GKK279 pKa = 9.82LAEE282 pKa = 4.08FRR284 pKa = 11.84AANPEE289 pKa = 3.91FFLCEE294 pKa = 4.08VKK296 pKa = 10.69NKK298 pKa = 9.43RR299 pKa = 11.84DD300 pKa = 3.38RR301 pKa = 11.84YY302 pKa = 7.66EE303 pKa = 3.79TAKK306 pKa = 9.15ITEE309 pKa = 3.96KK310 pKa = 9.91TRR312 pKa = 11.84PYY314 pKa = 10.68VAFPAHH320 pKa = 5.85FQLLFSCLSQPFTKK334 pKa = 10.48DD335 pKa = 2.7LLLFHH340 pKa = 6.84EE341 pKa = 5.23HH342 pKa = 6.6PKK344 pKa = 10.61SVNAYY349 pKa = 8.77GFTMAHH355 pKa = 6.37GGSSKK360 pKa = 9.92IRR362 pKa = 11.84YY363 pKa = 6.49RR364 pKa = 11.84TAEE367 pKa = 3.86LRR369 pKa = 11.84EE370 pKa = 4.26LGSKK374 pKa = 10.29GGAPICRR381 pKa = 11.84HH382 pKa = 5.96YY383 pKa = 11.41GDD385 pKa = 5.16DD386 pKa = 3.23ADD388 pKa = 3.55IYY390 pKa = 10.24VRR392 pKa = 11.84RR393 pKa = 11.84GGKK396 pKa = 8.88LLRR399 pKa = 11.84LCPDD403 pKa = 4.16FSQMDD408 pKa = 3.69GSVTHH413 pKa = 5.4EE414 pKa = 4.24TVRR417 pKa = 11.84LTVDD421 pKa = 4.06WILASYY427 pKa = 9.26EE428 pKa = 4.29EE429 pKa = 4.21QWGPSDD435 pKa = 3.66FWRR438 pKa = 11.84TIGDD442 pKa = 3.72LWVEE446 pKa = 4.32MATSPLFVVHH456 pKa = 7.08GKK458 pKa = 9.67DD459 pKa = 3.04VWEE462 pKa = 4.81KK463 pKa = 9.12RR464 pKa = 11.84TQDD467 pKa = 3.32GLMTGVVGTTLFDD480 pKa = 3.66TVHH483 pKa = 6.69AALAYY488 pKa = 7.21WVWVEE493 pKa = 3.78RR494 pKa = 11.84VHH496 pKa = 6.29YY497 pKa = 9.28QRR499 pKa = 11.84QYY501 pKa = 11.88ALLEE505 pKa = 3.88EE506 pKa = 4.6APAIKK511 pKa = 10.18LFKK514 pKa = 10.98EE515 pKa = 3.7MGLVVKK521 pKa = 10.3PGTWGFEE528 pKa = 4.26TVNEE532 pKa = 4.13NPVQGTLWTTQRR544 pKa = 11.84FLGVQYY550 pKa = 10.59KK551 pKa = 9.02WEE553 pKa = 4.17TGPRR557 pKa = 11.84EE558 pKa = 4.42VEE560 pKa = 4.33MVPHH564 pKa = 6.92LAYY567 pKa = 10.18EE568 pKa = 4.32DD569 pKa = 3.78WLSLLLCPRR578 pKa = 11.84GDD580 pKa = 3.34EE581 pKa = 4.41DD582 pKa = 3.61MRR584 pKa = 11.84SSRR587 pKa = 11.84TSEE590 pKa = 3.76LTRR593 pKa = 11.84EE594 pKa = 3.95RR595 pKa = 11.84VRR597 pKa = 11.84FDD599 pKa = 3.34RR600 pKa = 11.84LRR602 pKa = 11.84GMIVTGAFSTPEE614 pKa = 3.72ALAIMYY620 pKa = 8.39EE621 pKa = 4.55LIDD624 pKa = 3.9EE625 pKa = 4.57VNPVAIIMDD634 pKa = 3.86VQSGDD639 pKa = 3.74GKK641 pKa = 11.19GVTPDD646 pKa = 4.74AIALLNEE653 pKa = 4.27DD654 pKa = 4.2FQWPGSEE661 pKa = 5.62GVPDD665 pKa = 4.31WKK667 pKa = 10.46WCMNLYY673 pKa = 10.44LSPEE677 pKa = 3.98NQYY680 pKa = 11.39EE681 pKa = 3.97DD682 pKa = 4.4AQWKK686 pKa = 9.45LVIPEE691 pKa = 3.95AHH693 pKa = 6.74AALSDD698 pKa = 3.43FKK700 pKa = 11.41VRR702 pKa = 11.84YY703 pKa = 9.79RR704 pKa = 11.84KK705 pKa = 9.56DD706 pKa = 3.35LKK708 pKa = 11.04KK709 pKa = 10.57LATVQKK715 pKa = 10.11DD716 pKa = 3.81VEE718 pKa = 4.56GKK720 pKa = 9.48PSAAAMLPIPDD731 pKa = 5.16PAPKK735 pKa = 10.26AQIPSMEE742 pKa = 4.27IPTSGSRR749 pKa = 11.84VPKK752 pKa = 10.17TSEE755 pKa = 4.11AYY757 pKa = 10.49NKK759 pKa = 6.78TTKK762 pKa = 10.39PVDD765 pKa = 3.41VTLDD769 pKa = 3.63DD770 pKa = 3.25QGLAVEE776 pKa = 4.42EE777 pKa = 4.29RR778 pKa = 11.84TRR780 pKa = 11.84RR781 pKa = 11.84PTDD784 pKa = 3.12VEE786 pKa = 4.32QILSTFRR793 pKa = 11.84DD794 pKa = 3.72LPLSPKK800 pKa = 9.78AWEE803 pKa = 4.55EE804 pKa = 3.83IVAGATEE811 pKa = 4.12TQRR814 pKa = 11.84AGLEE818 pKa = 3.96ALQGGTKK825 pKa = 9.7VVQPVFQASTLSWKK839 pKa = 10.71LNWTVSKK846 pKa = 10.89VLSTAQKK853 pKa = 10.03AGLVVLPGGPKK864 pKa = 9.8RR865 pKa = 11.84KK866 pKa = 9.87SLITALPPSDD876 pKa = 4.1PSALEE881 pKa = 4.91PIDD884 pKa = 3.73HH885 pKa = 6.92AAAAVEE891 pKa = 3.99RR892 pKa = 11.84ALDD895 pKa = 3.63NSRR898 pKa = 11.84PEE900 pKa = 3.82PKK902 pKa = 9.32EE903 pKa = 3.32WSRR906 pKa = 11.84EE907 pKa = 3.86HH908 pKa = 6.21SQLGLSRR915 pKa = 11.84LKK917 pKa = 11.13GPLTQEE923 pKa = 3.84QAAKK927 pKa = 9.81VCKK930 pKa = 9.94EE931 pKa = 3.84FLAHH935 pKa = 7.78GYY937 pKa = 8.73RR938 pKa = 11.84CPMPKK943 pKa = 10.1CKK945 pKa = 10.02FGKK948 pKa = 10.51GNAAPLTFVEE958 pKa = 5.32VTLHH962 pKa = 6.98WITNDD967 pKa = 3.14PHH969 pKa = 7.38QDD971 pKa = 2.81KK972 pKa = 11.0CLLSKK977 pKa = 11.28CEE979 pKa = 4.22LHH981 pKa = 7.21DD982 pKa = 4.28GCPTGPPMQLRR993 pKa = 11.84PAKK996 pKa = 10.24EE997 pKa = 3.29ITLLKK1002 pKa = 10.16KK1003 pKa = 9.9QAEE1006 pKa = 4.31RR1007 pKa = 11.84LDD1009 pKa = 4.11GPQAEE1014 pKa = 4.69VLPPYY1019 pKa = 9.41PRR1021 pKa = 11.84PPLKK1025 pKa = 10.3FSSMDD1030 pKa = 3.35LEE1032 pKa = 4.77EE1033 pKa = 4.28IRR1035 pKa = 11.84SIQAEE1040 pKa = 4.58DD1041 pKa = 3.53PSMARR1046 pKa = 11.84SVFSSMCHH1054 pKa = 4.34EE1055 pKa = 4.18QGVAVVFTTKK1065 pKa = 9.83GTDD1068 pKa = 3.31HH1069 pKa = 7.07EE1070 pKa = 4.76KK1071 pKa = 10.98GAVNAPTYY1079 pKa = 11.39GEE1081 pKa = 4.16MHH1083 pKa = 6.14WTRR1086 pKa = 11.84IKK1088 pKa = 10.82KK1089 pKa = 9.65QGQPTPSFKK1098 pKa = 10.58RR1099 pKa = 11.84EE1100 pKa = 3.59PLAWNLYY1107 pKa = 9.47LQVLGKK1113 pKa = 10.17KK1114 pKa = 9.7RR1115 pKa = 11.84EE1116 pKa = 4.04LHH1118 pKa = 5.96NVLLTTALASLSDD1131 pKa = 3.89LGKK1134 pKa = 9.88PSLPPASWYY1143 pKa = 9.24EE1144 pKa = 3.8AAKK1147 pKa = 10.96VEE1149 pKa = 4.4LLPTASKK1156 pKa = 10.63AYY1158 pKa = 9.26STVKK1162 pKa = 10.08TPAPVLLNSKK1172 pKa = 9.65PSKK1175 pKa = 9.79TPLSQSISHH1184 pKa = 7.0AYY1186 pKa = 6.34EE1187 pKa = 4.05TKK1189 pKa = 10.08PQPSKK1194 pKa = 10.86PKK1196 pKa = 9.68TEE1198 pKa = 4.39SKK1200 pKa = 10.51PISKK1204 pKa = 9.06WGKK1207 pKa = 9.37KK1208 pKa = 9.48KK1209 pKa = 9.88PAAPGQAKK1217 pKa = 9.75KK1218 pKa = 10.72KK1219 pKa = 9.97KK1220 pKa = 9.02GPKK1223 pKa = 8.58KK1224 pKa = 9.65PSYY1227 pKa = 9.91GRR1229 pKa = 11.84SGATGSLPNRR1239 pKa = 11.84NSQQ1242 pKa = 3.15
Molecular weight: 138.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KI24|A0A1L3KI24_9VIRU Uncharacterized protein OS=Sanxia water strider virus 19 OX=1923403 PE=4 SV=1
MM1 pKa = 6.52PTKK4 pKa = 10.55RR5 pKa = 11.84SRR7 pKa = 11.84NPRR10 pKa = 11.84NPKK13 pKa = 9.61PNPNQSQNGAKK24 pKa = 10.23RR25 pKa = 11.84NPQPRR30 pKa = 11.84ARR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GPRR40 pKa = 11.84SPPMAGVAQQAAFQTGTRR58 pKa = 11.84NDD60 pKa = 4.16HH61 pKa = 6.2IRR63 pKa = 11.84VTGTDD68 pKa = 3.08KK69 pKa = 11.32LYY71 pKa = 11.43GEE73 pKa = 5.34DD74 pKa = 4.12NVPQKK79 pKa = 10.92HH80 pKa = 5.83RR81 pKa = 11.84GEE83 pKa = 4.03VLLAIRR89 pKa = 11.84INPASFPRR97 pKa = 11.84LSRR100 pKa = 11.84MASTFQKK107 pKa = 10.68YY108 pKa = 8.61RR109 pKa = 11.84FLRR112 pKa = 11.84MDD114 pKa = 3.54FRR116 pKa = 11.84IVSMASTSTSGGYY129 pKa = 9.96IIGFIPDD136 pKa = 4.31PDD138 pKa = 4.33DD139 pKa = 5.94LADD142 pKa = 3.24TWTTEE147 pKa = 3.92QLLSTPHH154 pKa = 5.68CRR156 pKa = 11.84IVKK159 pKa = 8.96AWEE162 pKa = 3.92SATIGATLEE171 pKa = 4.44SKK173 pKa = 9.38WLYY176 pKa = 9.9TNTGMEE182 pKa = 4.16SRR184 pKa = 11.84LYY186 pKa = 10.66SPGIFVIMVEE196 pKa = 4.31GSIPGTAPLSVYY208 pKa = 10.16VDD210 pKa = 3.82WVAEE214 pKa = 3.87MHH216 pKa = 6.15TPSLEE221 pKa = 4.2LVSKK225 pKa = 9.77PVQQIEE231 pKa = 4.59VVKK234 pKa = 10.68NAYY237 pKa = 10.1LRR239 pKa = 11.84DD240 pKa = 3.42AHH242 pKa = 6.36FGLWFSDD249 pKa = 3.59AEE251 pKa = 4.55GGDD254 pKa = 4.08DD255 pKa = 3.7PHH257 pKa = 7.34TIVPGIVYY265 pKa = 10.1DD266 pKa = 3.8VVYY269 pKa = 10.57KK270 pKa = 10.43LQSRR274 pKa = 11.84VYY276 pKa = 11.01VDD278 pKa = 3.5YY279 pKa = 10.76TDD281 pKa = 3.69QAGSYY286 pKa = 9.78DD287 pKa = 4.33KK288 pKa = 10.9IVATTGDD295 pKa = 3.5SHH297 pKa = 7.41GPTLWVADD305 pKa = 4.21FKK307 pKa = 11.33GRR309 pKa = 11.84RR310 pKa = 11.84IEE312 pKa = 4.26KK313 pKa = 10.16EE314 pKa = 3.67PVKK317 pKa = 10.45NTFVLEE323 pKa = 4.35KK324 pKa = 10.63GDD326 pKa = 3.49VLTPVQGEE334 pKa = 4.2VRR336 pKa = 11.84RR337 pKa = 11.84GTSCFPRR344 pKa = 11.84SPLPSSQDD352 pKa = 2.87GWEE355 pKa = 4.28LLQKK359 pKa = 10.24DD360 pKa = 4.43LRR362 pKa = 11.84NLSIEE367 pKa = 4.41GSSKK371 pKa = 11.01
MM1 pKa = 6.52PTKK4 pKa = 10.55RR5 pKa = 11.84SRR7 pKa = 11.84NPRR10 pKa = 11.84NPKK13 pKa = 9.61PNPNQSQNGAKK24 pKa = 10.23RR25 pKa = 11.84NPQPRR30 pKa = 11.84ARR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GPRR40 pKa = 11.84SPPMAGVAQQAAFQTGTRR58 pKa = 11.84NDD60 pKa = 4.16HH61 pKa = 6.2IRR63 pKa = 11.84VTGTDD68 pKa = 3.08KK69 pKa = 11.32LYY71 pKa = 11.43GEE73 pKa = 5.34DD74 pKa = 4.12NVPQKK79 pKa = 10.92HH80 pKa = 5.83RR81 pKa = 11.84GEE83 pKa = 4.03VLLAIRR89 pKa = 11.84INPASFPRR97 pKa = 11.84LSRR100 pKa = 11.84MASTFQKK107 pKa = 10.68YY108 pKa = 8.61RR109 pKa = 11.84FLRR112 pKa = 11.84MDD114 pKa = 3.54FRR116 pKa = 11.84IVSMASTSTSGGYY129 pKa = 9.96IIGFIPDD136 pKa = 4.31PDD138 pKa = 4.33DD139 pKa = 5.94LADD142 pKa = 3.24TWTTEE147 pKa = 3.92QLLSTPHH154 pKa = 5.68CRR156 pKa = 11.84IVKK159 pKa = 8.96AWEE162 pKa = 3.92SATIGATLEE171 pKa = 4.44SKK173 pKa = 9.38WLYY176 pKa = 9.9TNTGMEE182 pKa = 4.16SRR184 pKa = 11.84LYY186 pKa = 10.66SPGIFVIMVEE196 pKa = 4.31GSIPGTAPLSVYY208 pKa = 10.16VDD210 pKa = 3.82WVAEE214 pKa = 3.87MHH216 pKa = 6.15TPSLEE221 pKa = 4.2LVSKK225 pKa = 9.77PVQQIEE231 pKa = 4.59VVKK234 pKa = 10.68NAYY237 pKa = 10.1LRR239 pKa = 11.84DD240 pKa = 3.42AHH242 pKa = 6.36FGLWFSDD249 pKa = 3.59AEE251 pKa = 4.55GGDD254 pKa = 4.08DD255 pKa = 3.7PHH257 pKa = 7.34TIVPGIVYY265 pKa = 10.1DD266 pKa = 3.8VVYY269 pKa = 10.57KK270 pKa = 10.43LQSRR274 pKa = 11.84VYY276 pKa = 11.01VDD278 pKa = 3.5YY279 pKa = 10.76TDD281 pKa = 3.69QAGSYY286 pKa = 9.78DD287 pKa = 4.33KK288 pKa = 10.9IVATTGDD295 pKa = 3.5SHH297 pKa = 7.41GPTLWVADD305 pKa = 4.21FKK307 pKa = 11.33GRR309 pKa = 11.84RR310 pKa = 11.84IEE312 pKa = 4.26KK313 pKa = 10.16EE314 pKa = 3.67PVKK317 pKa = 10.45NTFVLEE323 pKa = 4.35KK324 pKa = 10.63GDD326 pKa = 3.49VLTPVQGEE334 pKa = 4.2VRR336 pKa = 11.84RR337 pKa = 11.84GTSCFPRR344 pKa = 11.84SPLPSSQDD352 pKa = 2.87GWEE355 pKa = 4.28LLQKK359 pKa = 10.24DD360 pKa = 4.43LRR362 pKa = 11.84NLSIEE367 pKa = 4.41GSSKK371 pKa = 11.01
Molecular weight: 41.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1733 |
120 |
1242 |
577.7 |
64.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.501 ± 0.694 | 1.212 ± 0.3 |
5.309 ± 0.647 | 6.29 ± 0.703 |
3.001 ± 0.135 | 6.52 ± 1.231 |
2.308 ± 0.16 | 3.347 ± 0.586 |
6.694 ± 1.063 | 9.521 ± 1.517 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.366 ± 0.209 | 2.25 ± 0.646 |
8.194 ± 0.635 | 4.328 ± 0.165 |
6.636 ± 1.37 | 7.271 ± 0.317 |
6.174 ± 1.284 | 6.463 ± 0.788 |
1.962 ± 0.289 | 2.654 ± 0.453 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
