
Spirochaeta africana (strain ATCC 700263 / DSM 8902 / Z-7692)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Spirochaetaceae; Spirochaeta; Spirochaeta africana
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
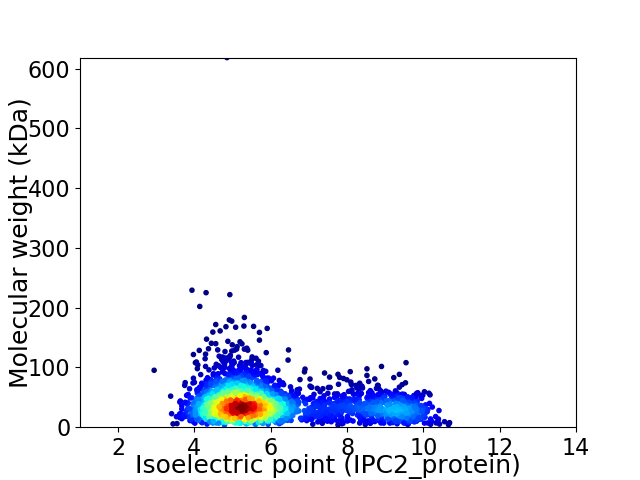
Virtual 2D-PAGE plot for 2766 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H9UG03|H9UG03_SPIAZ Putative ATPase of the PP-loop superfamily implicated in cell cycle control OS=Spirochaeta africana (strain ATCC 700263 / DSM 8902 / Z-7692) OX=889378 GN=Spiaf_0340 PE=4 SV=1
MM1 pKa = 6.77MWKK4 pKa = 9.78SLKK7 pKa = 10.44SSFVGVLLVLCIVVSAGSCSNLFEE31 pKa = 6.21AGEE34 pKa = 4.24RR35 pKa = 11.84TVALQLEE42 pKa = 4.76VRR44 pKa = 11.84APALFPAADD53 pKa = 3.35ARR55 pKa = 11.84GRR57 pKa = 11.84TILPDD62 pKa = 3.05AAMAEE67 pKa = 4.57VVLLEE72 pKa = 4.42HH73 pKa = 6.89NGASLQQHH81 pKa = 6.79HH82 pKa = 7.18LPLTYY87 pKa = 10.69DD88 pKa = 3.34PVLQEE93 pKa = 4.15FHH95 pKa = 6.62ATLLLEE101 pKa = 4.99DD102 pKa = 3.98VPAGNSHH109 pKa = 6.58ILAVEE114 pKa = 3.99IQDD117 pKa = 3.95EE118 pKa = 4.44EE119 pKa = 4.53GAPLQEE125 pKa = 4.18AQAQVSVVSGEE136 pKa = 4.32TTSVTLILFPHH147 pKa = 6.9TDD149 pKa = 3.42LVQPGDD155 pKa = 3.49LAEE158 pKa = 4.85DD159 pKa = 3.86MEE161 pKa = 4.77EE162 pKa = 4.11TVPPGGYY169 pKa = 9.23WILAYY174 pKa = 10.59SLAAGQDD181 pKa = 3.63YY182 pKa = 11.07ALEE185 pKa = 4.7AEE187 pKa = 4.28AGEE190 pKa = 4.79DD191 pKa = 3.12IEE193 pKa = 6.37LRR195 pKa = 11.84IQLPDD200 pKa = 3.52GSFLPAGEE208 pKa = 4.33ITWEE212 pKa = 4.03QQEE215 pKa = 4.37TGTVYY220 pKa = 10.84LYY222 pKa = 10.49IFNRR226 pKa = 11.84GDD228 pKa = 3.35DD229 pKa = 4.98DD230 pKa = 4.64IPVLLEE236 pKa = 3.8LTASGAVEE244 pKa = 3.88YY245 pKa = 9.9TIGDD249 pKa = 3.62TGPAGGVVFYY259 pKa = 11.0DD260 pKa = 2.95QGEE263 pKa = 4.57VVNGWRR269 pKa = 11.84YY270 pKa = 10.6LEE272 pKa = 4.19ATPAGWSGGEE282 pKa = 4.28DD283 pKa = 3.29PVAAWGDD290 pKa = 3.49DD291 pKa = 3.57SLIGTEE297 pKa = 4.29TNFGFGKK304 pKa = 10.59LNTLTIISRR313 pKa = 11.84LQDD316 pKa = 3.26WEE318 pKa = 4.41DD319 pKa = 3.26GQYY322 pKa = 11.13AARR325 pKa = 11.84LSGDD329 pKa = 3.86YY330 pKa = 10.96AVGEE334 pKa = 4.07YY335 pKa = 10.64DD336 pKa = 4.88DD337 pKa = 4.41WFLPSLEE344 pKa = 4.29EE345 pKa = 4.17LFEE348 pKa = 4.98LRR350 pKa = 11.84AQQDD354 pKa = 3.29IVGGFPIEE362 pKa = 4.24GNEE365 pKa = 4.07DD366 pKa = 3.84IYY368 pKa = 10.89WSSSEE373 pKa = 5.04FFDD376 pKa = 6.22DD377 pKa = 4.86PDD379 pKa = 4.55DD380 pKa = 4.5PPEE383 pKa = 3.84EE384 pKa = 4.38HH385 pKa = 6.72SWVLGFIAGEE395 pKa = 3.83SSEE398 pKa = 4.02WLKK401 pKa = 11.1EE402 pKa = 3.87FDD404 pKa = 3.83DD405 pKa = 3.9TRR407 pKa = 11.84VRR409 pKa = 11.84PVRR412 pKa = 11.84AFRR415 pKa = 11.84SEE417 pKa = 4.05LPTYY421 pKa = 10.5AVVYY425 pKa = 9.91HH426 pKa = 6.42PNEE429 pKa = 4.37GQGDD433 pKa = 4.12PPVDD437 pKa = 4.0PYY439 pKa = 10.88HH440 pKa = 6.61YY441 pKa = 10.48EE442 pKa = 3.82PGEE445 pKa = 4.19PVTLPDD451 pKa = 3.64AGEE454 pKa = 4.47LYY456 pKa = 10.57RR457 pKa = 11.84EE458 pKa = 4.8GYY460 pKa = 9.78EE461 pKa = 3.88FAGWNSEE468 pKa = 3.85PDD470 pKa = 3.57GTGDD474 pKa = 3.42QYY476 pKa = 11.61IAIGNITMPQGNLILYY492 pKa = 9.46AEE494 pKa = 4.29WAEE497 pKa = 3.94IEE499 pKa = 4.07PVYY502 pKa = 10.52TIGDD506 pKa = 3.63TGPAGGIIFYY516 pKa = 10.95VDD518 pKa = 3.96DD519 pKa = 3.72TDD521 pKa = 5.82QYY523 pKa = 12.05DD524 pKa = 2.82GWTYY528 pKa = 11.42LEE530 pKa = 4.62AAPAGWDD537 pKa = 3.85DD538 pKa = 4.25PGEE541 pKa = 4.49PDD543 pKa = 4.38PEE545 pKa = 5.68DD546 pKa = 5.02PLRR549 pKa = 11.84QWDD552 pKa = 4.66DD553 pKa = 3.33DD554 pKa = 4.1TFSYY558 pKa = 10.85SAVTTEE564 pKa = 4.03TAIGTGKK571 pKa = 10.32EE572 pKa = 3.55NTAAIVTALGGDD584 pKa = 4.09GKK586 pKa = 9.38TGMAAQLAAEE596 pKa = 4.3LEE598 pKa = 4.38ITNGEE603 pKa = 4.03TDD605 pKa = 3.38YY606 pKa = 11.79SDD608 pKa = 3.01WFLPSRR614 pKa = 11.84DD615 pKa = 4.12EE616 pKa = 4.59LNEE619 pKa = 4.3LYY621 pKa = 9.34MQEE624 pKa = 4.44AIIDD628 pKa = 3.91GLYY631 pKa = 10.73VSAGWYY637 pKa = 8.41YY638 pKa = 10.14WSSSTADD645 pKa = 3.91EE646 pKa = 4.12SWAWSQDD653 pKa = 3.22FFNGDD658 pKa = 3.27QSVTNGKK665 pKa = 9.73NISTYY670 pKa = 10.08IRR672 pKa = 11.84VRR674 pKa = 11.84PIRR677 pKa = 11.84RR678 pKa = 11.84FF679 pKa = 3.22
MM1 pKa = 6.77MWKK4 pKa = 9.78SLKK7 pKa = 10.44SSFVGVLLVLCIVVSAGSCSNLFEE31 pKa = 6.21AGEE34 pKa = 4.24RR35 pKa = 11.84TVALQLEE42 pKa = 4.76VRR44 pKa = 11.84APALFPAADD53 pKa = 3.35ARR55 pKa = 11.84GRR57 pKa = 11.84TILPDD62 pKa = 3.05AAMAEE67 pKa = 4.57VVLLEE72 pKa = 4.42HH73 pKa = 6.89NGASLQQHH81 pKa = 6.79HH82 pKa = 7.18LPLTYY87 pKa = 10.69DD88 pKa = 3.34PVLQEE93 pKa = 4.15FHH95 pKa = 6.62ATLLLEE101 pKa = 4.99DD102 pKa = 3.98VPAGNSHH109 pKa = 6.58ILAVEE114 pKa = 3.99IQDD117 pKa = 3.95EE118 pKa = 4.44EE119 pKa = 4.53GAPLQEE125 pKa = 4.18AQAQVSVVSGEE136 pKa = 4.32TTSVTLILFPHH147 pKa = 6.9TDD149 pKa = 3.42LVQPGDD155 pKa = 3.49LAEE158 pKa = 4.85DD159 pKa = 3.86MEE161 pKa = 4.77EE162 pKa = 4.11TVPPGGYY169 pKa = 9.23WILAYY174 pKa = 10.59SLAAGQDD181 pKa = 3.63YY182 pKa = 11.07ALEE185 pKa = 4.7AEE187 pKa = 4.28AGEE190 pKa = 4.79DD191 pKa = 3.12IEE193 pKa = 6.37LRR195 pKa = 11.84IQLPDD200 pKa = 3.52GSFLPAGEE208 pKa = 4.33ITWEE212 pKa = 4.03QQEE215 pKa = 4.37TGTVYY220 pKa = 10.84LYY222 pKa = 10.49IFNRR226 pKa = 11.84GDD228 pKa = 3.35DD229 pKa = 4.98DD230 pKa = 4.64IPVLLEE236 pKa = 3.8LTASGAVEE244 pKa = 3.88YY245 pKa = 9.9TIGDD249 pKa = 3.62TGPAGGVVFYY259 pKa = 11.0DD260 pKa = 2.95QGEE263 pKa = 4.57VVNGWRR269 pKa = 11.84YY270 pKa = 10.6LEE272 pKa = 4.19ATPAGWSGGEE282 pKa = 4.28DD283 pKa = 3.29PVAAWGDD290 pKa = 3.49DD291 pKa = 3.57SLIGTEE297 pKa = 4.29TNFGFGKK304 pKa = 10.59LNTLTIISRR313 pKa = 11.84LQDD316 pKa = 3.26WEE318 pKa = 4.41DD319 pKa = 3.26GQYY322 pKa = 11.13AARR325 pKa = 11.84LSGDD329 pKa = 3.86YY330 pKa = 10.96AVGEE334 pKa = 4.07YY335 pKa = 10.64DD336 pKa = 4.88DD337 pKa = 4.41WFLPSLEE344 pKa = 4.29EE345 pKa = 4.17LFEE348 pKa = 4.98LRR350 pKa = 11.84AQQDD354 pKa = 3.29IVGGFPIEE362 pKa = 4.24GNEE365 pKa = 4.07DD366 pKa = 3.84IYY368 pKa = 10.89WSSSEE373 pKa = 5.04FFDD376 pKa = 6.22DD377 pKa = 4.86PDD379 pKa = 4.55DD380 pKa = 4.5PPEE383 pKa = 3.84EE384 pKa = 4.38HH385 pKa = 6.72SWVLGFIAGEE395 pKa = 3.83SSEE398 pKa = 4.02WLKK401 pKa = 11.1EE402 pKa = 3.87FDD404 pKa = 3.83DD405 pKa = 3.9TRR407 pKa = 11.84VRR409 pKa = 11.84PVRR412 pKa = 11.84AFRR415 pKa = 11.84SEE417 pKa = 4.05LPTYY421 pKa = 10.5AVVYY425 pKa = 9.91HH426 pKa = 6.42PNEE429 pKa = 4.37GQGDD433 pKa = 4.12PPVDD437 pKa = 4.0PYY439 pKa = 10.88HH440 pKa = 6.61YY441 pKa = 10.48EE442 pKa = 3.82PGEE445 pKa = 4.19PVTLPDD451 pKa = 3.64AGEE454 pKa = 4.47LYY456 pKa = 10.57RR457 pKa = 11.84EE458 pKa = 4.8GYY460 pKa = 9.78EE461 pKa = 3.88FAGWNSEE468 pKa = 3.85PDD470 pKa = 3.57GTGDD474 pKa = 3.42QYY476 pKa = 11.61IAIGNITMPQGNLILYY492 pKa = 9.46AEE494 pKa = 4.29WAEE497 pKa = 3.94IEE499 pKa = 4.07PVYY502 pKa = 10.52TIGDD506 pKa = 3.63TGPAGGIIFYY516 pKa = 10.95VDD518 pKa = 3.96DD519 pKa = 3.72TDD521 pKa = 5.82QYY523 pKa = 12.05DD524 pKa = 2.82GWTYY528 pKa = 11.42LEE530 pKa = 4.62AAPAGWDD537 pKa = 3.85DD538 pKa = 4.25PGEE541 pKa = 4.49PDD543 pKa = 4.38PEE545 pKa = 5.68DD546 pKa = 5.02PLRR549 pKa = 11.84QWDD552 pKa = 4.66DD553 pKa = 3.33DD554 pKa = 4.1TFSYY558 pKa = 10.85SAVTTEE564 pKa = 4.03TAIGTGKK571 pKa = 10.32EE572 pKa = 3.55NTAAIVTALGGDD584 pKa = 4.09GKK586 pKa = 9.38TGMAAQLAAEE596 pKa = 4.3LEE598 pKa = 4.38ITNGEE603 pKa = 4.03TDD605 pKa = 3.38YY606 pKa = 11.79SDD608 pKa = 3.01WFLPSRR614 pKa = 11.84DD615 pKa = 4.12EE616 pKa = 4.59LNEE619 pKa = 4.3LYY621 pKa = 9.34MQEE624 pKa = 4.44AIIDD628 pKa = 3.91GLYY631 pKa = 10.73VSAGWYY637 pKa = 8.41YY638 pKa = 10.14WSSSTADD645 pKa = 3.91EE646 pKa = 4.12SWAWSQDD653 pKa = 3.22FFNGDD658 pKa = 3.27QSVTNGKK665 pKa = 9.73NISTYY670 pKa = 10.08IRR672 pKa = 11.84VRR674 pKa = 11.84PIRR677 pKa = 11.84RR678 pKa = 11.84FF679 pKa = 3.22
Molecular weight: 74.57 kDa
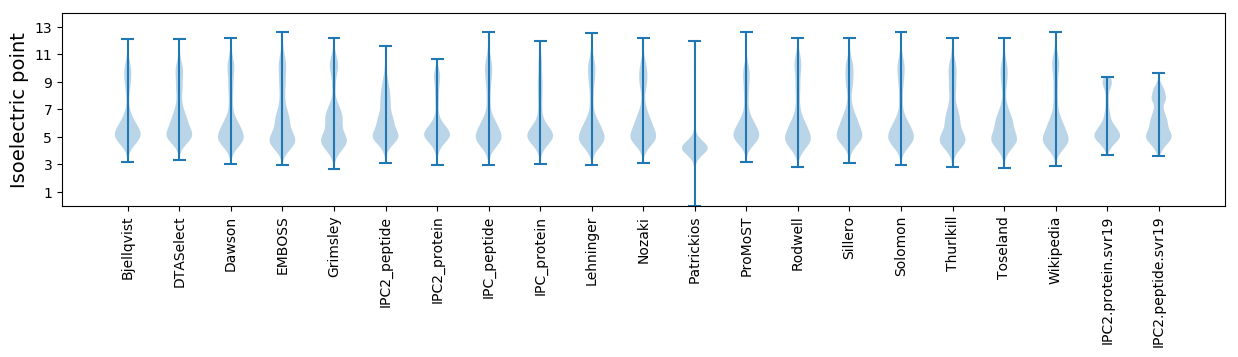
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H9UMJ8|H9UMJ8_SPIAZ DNA polymerase I OS=Spirochaeta africana (strain ATCC 700263 / DSM 8902 / Z-7692) OX=889378 GN=polA PE=3 SV=1
MM1 pKa = 7.18QLRR4 pKa = 11.84DD5 pKa = 3.52IVTPLIILPAAAVLSWVFPQYY26 pKa = 9.36NTAFIQVGLQFVALLFFLAGILLSILYY53 pKa = 10.1HH54 pKa = 6.55RR55 pKa = 11.84GRR57 pKa = 11.84VLAALMLMLLTAAAFASPVVLLLEE81 pKa = 5.16PYY83 pKa = 8.23QTTMVYY89 pKa = 10.5RR90 pKa = 11.84LLQLSMPLLLCVIAISPRR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84ILDD113 pKa = 3.66PASLVWLASIALLVFAVWAVPNTEE137 pKa = 3.77QAPRR141 pKa = 11.84LLQPLFTLQLLPFLPPQLLPQALPQISIAMALFGLLCVILVEE183 pKa = 4.56STYY186 pKa = 10.88HH187 pKa = 6.88RR188 pKa = 11.84DD189 pKa = 3.49PSLSLAWICAYY200 pKa = 10.47AAVCLALITLEE211 pKa = 4.31TPARR215 pKa = 11.84AILWFTAAGAVLAVGLIQHH234 pKa = 6.49AHH236 pKa = 5.66RR237 pKa = 11.84MAFIDD242 pKa = 3.75ALTRR246 pKa = 11.84IPNRR250 pKa = 11.84RR251 pKa = 11.84ALDD254 pKa = 3.64EE255 pKa = 3.93QLSRR259 pKa = 11.84LRR261 pKa = 11.84SRR263 pKa = 11.84YY264 pKa = 9.43AIAMVDD270 pKa = 2.93IDD272 pKa = 4.29HH273 pKa = 7.03FKK275 pKa = 11.22AFNDD279 pKa = 3.67THH281 pKa = 4.85GHH283 pKa = 5.73KK284 pKa = 10.19MGDD287 pKa = 3.0QALRR291 pKa = 11.84KK292 pKa = 6.83TASHH296 pKa = 5.95LRR298 pKa = 11.84EE299 pKa = 3.58VDD301 pKa = 2.93GGGKK305 pKa = 8.01VFRR308 pKa = 11.84YY309 pKa = 9.89GGEE312 pKa = 3.8EE313 pKa = 3.79FTILFPGKK321 pKa = 6.75TARR324 pKa = 11.84EE325 pKa = 4.2AGRR328 pKa = 11.84HH329 pKa = 4.35CQALRR334 pKa = 11.84EE335 pKa = 4.33RR336 pKa = 11.84IAGEE340 pKa = 3.71PFFFRR345 pKa = 11.84GSNRR349 pKa = 11.84PRR351 pKa = 11.84KK352 pKa = 9.04LRR354 pKa = 11.84KK355 pKa = 9.28AEE357 pKa = 3.73KK358 pKa = 9.43HH359 pKa = 4.85RR360 pKa = 11.84RR361 pKa = 11.84RR362 pKa = 11.84PTRR365 pKa = 11.84KK366 pKa = 9.33RR367 pKa = 11.84EE368 pKa = 4.04ASDD371 pKa = 3.27SLQITVSIGVAAQGDD386 pKa = 4.02RR387 pKa = 11.84ASTSARR393 pKa = 11.84VLQLADD399 pKa = 3.17QALYY403 pKa = 9.82HH404 pKa = 6.25AKK406 pKa = 10.26KK407 pKa = 10.11NGRR410 pKa = 11.84NRR412 pKa = 11.84VIVARR417 pKa = 11.84DD418 pKa = 3.28DD419 pKa = 3.56APRR422 pKa = 5.18
MM1 pKa = 7.18QLRR4 pKa = 11.84DD5 pKa = 3.52IVTPLIILPAAAVLSWVFPQYY26 pKa = 9.36NTAFIQVGLQFVALLFFLAGILLSILYY53 pKa = 10.1HH54 pKa = 6.55RR55 pKa = 11.84GRR57 pKa = 11.84VLAALMLMLLTAAAFASPVVLLLEE81 pKa = 5.16PYY83 pKa = 8.23QTTMVYY89 pKa = 10.5RR90 pKa = 11.84LLQLSMPLLLCVIAISPRR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84ILDD113 pKa = 3.66PASLVWLASIALLVFAVWAVPNTEE137 pKa = 3.77QAPRR141 pKa = 11.84LLQPLFTLQLLPFLPPQLLPQALPQISIAMALFGLLCVILVEE183 pKa = 4.56STYY186 pKa = 10.88HH187 pKa = 6.88RR188 pKa = 11.84DD189 pKa = 3.49PSLSLAWICAYY200 pKa = 10.47AAVCLALITLEE211 pKa = 4.31TPARR215 pKa = 11.84AILWFTAAGAVLAVGLIQHH234 pKa = 6.49AHH236 pKa = 5.66RR237 pKa = 11.84MAFIDD242 pKa = 3.75ALTRR246 pKa = 11.84IPNRR250 pKa = 11.84RR251 pKa = 11.84ALDD254 pKa = 3.64EE255 pKa = 3.93QLSRR259 pKa = 11.84LRR261 pKa = 11.84SRR263 pKa = 11.84YY264 pKa = 9.43AIAMVDD270 pKa = 2.93IDD272 pKa = 4.29HH273 pKa = 7.03FKK275 pKa = 11.22AFNDD279 pKa = 3.67THH281 pKa = 4.85GHH283 pKa = 5.73KK284 pKa = 10.19MGDD287 pKa = 3.0QALRR291 pKa = 11.84KK292 pKa = 6.83TASHH296 pKa = 5.95LRR298 pKa = 11.84EE299 pKa = 3.58VDD301 pKa = 2.93GGGKK305 pKa = 8.01VFRR308 pKa = 11.84YY309 pKa = 9.89GGEE312 pKa = 3.8EE313 pKa = 3.79FTILFPGKK321 pKa = 6.75TARR324 pKa = 11.84EE325 pKa = 4.2AGRR328 pKa = 11.84HH329 pKa = 4.35CQALRR334 pKa = 11.84EE335 pKa = 4.33RR336 pKa = 11.84IAGEE340 pKa = 3.71PFFFRR345 pKa = 11.84GSNRR349 pKa = 11.84PRR351 pKa = 11.84KK352 pKa = 9.04LRR354 pKa = 11.84KK355 pKa = 9.28AEE357 pKa = 3.73KK358 pKa = 9.43HH359 pKa = 4.85RR360 pKa = 11.84RR361 pKa = 11.84RR362 pKa = 11.84PTRR365 pKa = 11.84KK366 pKa = 9.33RR367 pKa = 11.84EE368 pKa = 4.04ASDD371 pKa = 3.27SLQITVSIGVAAQGDD386 pKa = 4.02RR387 pKa = 11.84ASTSARR393 pKa = 11.84VLQLADD399 pKa = 3.17QALYY403 pKa = 9.82HH404 pKa = 6.25AKK406 pKa = 10.26KK407 pKa = 10.11NGRR410 pKa = 11.84NRR412 pKa = 11.84VIVARR417 pKa = 11.84DD418 pKa = 3.28DD419 pKa = 3.56APRR422 pKa = 5.18
Molecular weight: 46.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1014669 |
31 |
5749 |
366.8 |
40.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.707 ± 0.052 | 0.786 ± 0.015 |
5.795 ± 0.032 | 6.482 ± 0.045 |
3.862 ± 0.029 | 7.788 ± 0.049 |
2.308 ± 0.02 | 5.931 ± 0.042 |
2.638 ± 0.041 | 10.551 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.32 ± 0.02 | 2.856 ± 0.03 |
4.887 ± 0.037 | 4.533 ± 0.034 |
7.197 ± 0.042 | 6.054 ± 0.027 |
5.166 ± 0.032 | 6.92 ± 0.033 |
1.192 ± 0.019 | 3.027 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
