
Maize streak virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins
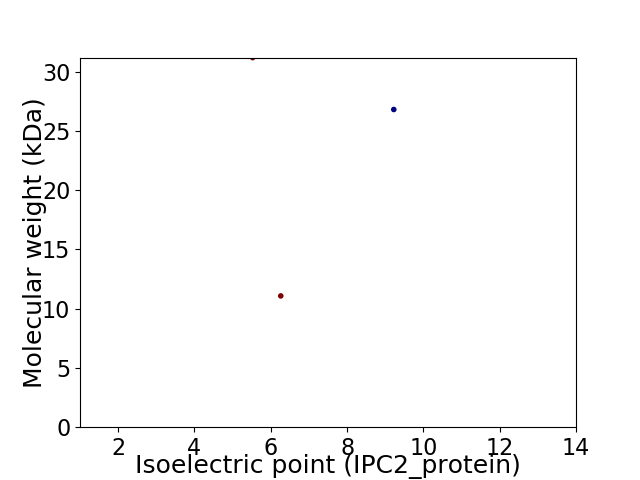
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
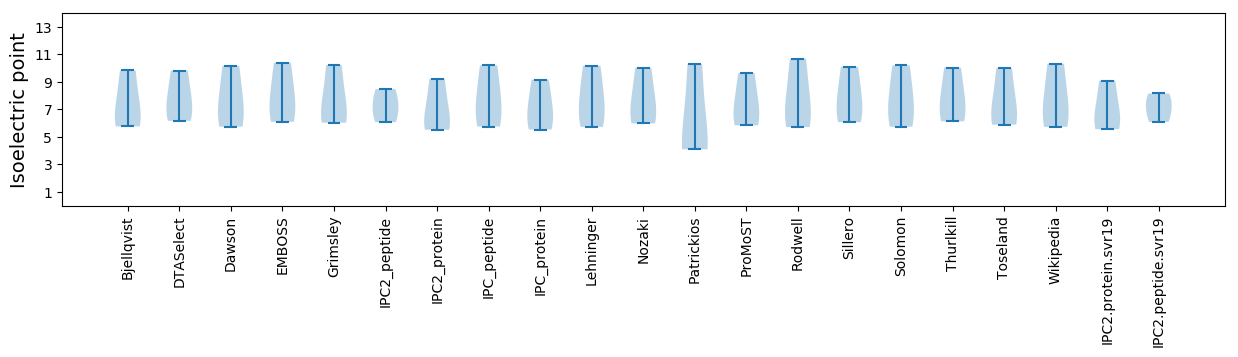
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
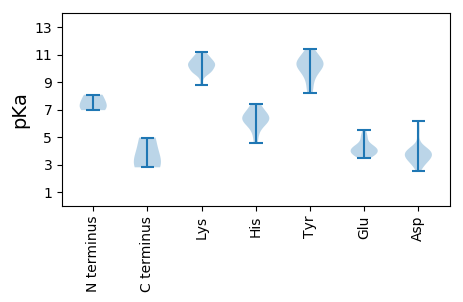
Protein with the lowest isoelectric point:
>tr|B6CZV2|B6CZV2_9GEMI Replication-associated protein OS=Maize streak virus OX=10821 PE=3 SV=1
MM1 pKa = 7.36TSSSSNRR8 pKa = 11.84QFLHH12 pKa = 6.69RR13 pKa = 11.84NVNTFLTYY21 pKa = 8.18PHH23 pKa = 7.01CPEE26 pKa = 3.84NPEE29 pKa = 4.12IVSQKK34 pKa = 9.79LWEE37 pKa = 4.12LVGRR41 pKa = 11.84WKK43 pKa = 10.25PLYY46 pKa = 9.81IICARR51 pKa = 11.84EE52 pKa = 3.67AHH54 pKa = 6.6EE55 pKa = 5.52DD56 pKa = 3.97GNLHH60 pKa = 6.05LHH62 pKa = 6.22VLIQTDD68 pKa = 3.77KK69 pKa = 10.81PVRR72 pKa = 11.84TSDD75 pKa = 3.81SRR77 pKa = 11.84FFDD80 pKa = 2.91IDD82 pKa = 3.48GFHH85 pKa = 7.27PNIQSAMSVNKK96 pKa = 9.81VRR98 pKa = 11.84DD99 pKa = 3.94YY100 pKa = 11.14ILKK103 pKa = 10.2EE104 pKa = 3.57PLALFEE110 pKa = 5.16RR111 pKa = 11.84GTFIPRR117 pKa = 11.84KK118 pKa = 9.66KK119 pKa = 10.36SVLGNSSKK127 pKa = 11.16GNSEE131 pKa = 4.12KK132 pKa = 10.7KK133 pKa = 9.55PSKK136 pKa = 10.67DD137 pKa = 3.89EE138 pKa = 3.83IMQDD142 pKa = 3.59IISHH146 pKa = 5.63ATSKK150 pKa = 10.82RR151 pKa = 11.84EE152 pKa = 3.77YY153 pKa = 10.24LSMVRR158 pKa = 11.84KK159 pKa = 9.97SFPYY163 pKa = 10.17DD164 pKa = 2.76WATKK168 pKa = 9.61LQYY171 pKa = 10.4FEE173 pKa = 4.89YY174 pKa = 10.06SASKK178 pKa = 10.37LFPDD182 pKa = 3.43IQEE185 pKa = 4.21EE186 pKa = 4.74FINPHH191 pKa = 6.03PTSEE195 pKa = 4.43PDD197 pKa = 3.53LLCNEE202 pKa = 5.12SIKK205 pKa = 11.07DD206 pKa = 3.33WLQPNIYY213 pKa = 9.41QVSPEE218 pKa = 4.47AYY220 pKa = 8.91MLLQPTCYY228 pKa = 10.31SVDD231 pKa = 3.65DD232 pKa = 5.6AISDD236 pKa = 4.18LQWLDD241 pKa = 3.79DD242 pKa = 4.03VSSKK246 pKa = 11.14QIMDD250 pKa = 3.34QGSRR254 pKa = 11.84ASTSSVQQGQGNLLGPEE271 pKa = 3.97AA272 pKa = 4.96
MM1 pKa = 7.36TSSSSNRR8 pKa = 11.84QFLHH12 pKa = 6.69RR13 pKa = 11.84NVNTFLTYY21 pKa = 8.18PHH23 pKa = 7.01CPEE26 pKa = 3.84NPEE29 pKa = 4.12IVSQKK34 pKa = 9.79LWEE37 pKa = 4.12LVGRR41 pKa = 11.84WKK43 pKa = 10.25PLYY46 pKa = 9.81IICARR51 pKa = 11.84EE52 pKa = 3.67AHH54 pKa = 6.6EE55 pKa = 5.52DD56 pKa = 3.97GNLHH60 pKa = 6.05LHH62 pKa = 6.22VLIQTDD68 pKa = 3.77KK69 pKa = 10.81PVRR72 pKa = 11.84TSDD75 pKa = 3.81SRR77 pKa = 11.84FFDD80 pKa = 2.91IDD82 pKa = 3.48GFHH85 pKa = 7.27PNIQSAMSVNKK96 pKa = 9.81VRR98 pKa = 11.84DD99 pKa = 3.94YY100 pKa = 11.14ILKK103 pKa = 10.2EE104 pKa = 3.57PLALFEE110 pKa = 5.16RR111 pKa = 11.84GTFIPRR117 pKa = 11.84KK118 pKa = 9.66KK119 pKa = 10.36SVLGNSSKK127 pKa = 11.16GNSEE131 pKa = 4.12KK132 pKa = 10.7KK133 pKa = 9.55PSKK136 pKa = 10.67DD137 pKa = 3.89EE138 pKa = 3.83IMQDD142 pKa = 3.59IISHH146 pKa = 5.63ATSKK150 pKa = 10.82RR151 pKa = 11.84EE152 pKa = 3.77YY153 pKa = 10.24LSMVRR158 pKa = 11.84KK159 pKa = 9.97SFPYY163 pKa = 10.17DD164 pKa = 2.76WATKK168 pKa = 9.61LQYY171 pKa = 10.4FEE173 pKa = 4.89YY174 pKa = 10.06SASKK178 pKa = 10.37LFPDD182 pKa = 3.43IQEE185 pKa = 4.21EE186 pKa = 4.74FINPHH191 pKa = 6.03PTSEE195 pKa = 4.43PDD197 pKa = 3.53LLCNEE202 pKa = 5.12SIKK205 pKa = 11.07DD206 pKa = 3.33WLQPNIYY213 pKa = 9.41QVSPEE218 pKa = 4.47AYY220 pKa = 8.91MLLQPTCYY228 pKa = 10.31SVDD231 pKa = 3.65DD232 pKa = 5.6AISDD236 pKa = 4.18LQWLDD241 pKa = 3.79DD242 pKa = 4.03VSSKK246 pKa = 11.14QIMDD250 pKa = 3.34QGSRR254 pKa = 11.84ASTSSVQQGQGNLLGPEE271 pKa = 3.97AA272 pKa = 4.96
Molecular weight: 31.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B6CZV0|B6CZV0_9GEMI Movement protein OS=Maize streak virus OX=10821 PE=3 SV=1
MM1 pKa = 6.95STSKK5 pKa = 10.63RR6 pKa = 11.84KK7 pKa = 9.69RR8 pKa = 11.84ADD10 pKa = 3.22EE11 pKa = 4.25AQWNQRR17 pKa = 11.84STKK20 pKa = 9.94KK21 pKa = 10.25KK22 pKa = 10.28ASAPKK27 pKa = 9.94AKK29 pKa = 10.2KK30 pKa = 10.35AGGKK34 pKa = 8.85GEE36 pKa = 4.18KK37 pKa = 9.62PSLQIQTLLHH47 pKa = 6.55SGDD50 pKa = 3.49TMITVPSGGVCDD62 pKa = 6.17LINTYY67 pKa = 11.02ARR69 pKa = 11.84GSDD72 pKa = 3.04EE73 pKa = 4.39GNRR76 pKa = 11.84HH77 pKa = 4.54TSEE80 pKa = 3.79TLTYY84 pKa = 10.21KK85 pKa = 10.8VGVDD89 pKa = 3.04YY90 pKa = 11.43HH91 pKa = 6.46FVADD95 pKa = 4.09AASCKK100 pKa = 9.91YY101 pKa = 10.2SNRR104 pKa = 11.84GTGVMWLVYY113 pKa = 9.39DD114 pKa = 4.05TTPGGNAPTTKK125 pKa = 10.51DD126 pKa = 2.52IFAYY130 pKa = 10.59SDD132 pKa = 3.75ALKK135 pKa = 10.5AWPTTWKK142 pKa = 10.31VSRR145 pKa = 11.84EE146 pKa = 3.85LCHH149 pKa = 6.49RR150 pKa = 11.84FVVKK154 pKa = 10.41RR155 pKa = 11.84RR156 pKa = 11.84WLFTMEE162 pKa = 3.72TDD164 pKa = 3.84GRR166 pKa = 11.84IGSDD170 pKa = 3.46TPPSNASWPPCKK182 pKa = 10.18RR183 pKa = 11.84NVDD186 pKa = 4.1FHH188 pKa = 7.43KK189 pKa = 9.79FTSGLGVRR197 pKa = 11.84TQWKK201 pKa = 8.76NVTDD205 pKa = 4.02GGVGAIQRR213 pKa = 11.84GALYY217 pKa = 10.5LVIAPGNGITFTAHH231 pKa = 5.19GQTRR235 pKa = 11.84LYY237 pKa = 9.73FKK239 pKa = 10.92SVGNQQ244 pKa = 2.8
MM1 pKa = 6.95STSKK5 pKa = 10.63RR6 pKa = 11.84KK7 pKa = 9.69RR8 pKa = 11.84ADD10 pKa = 3.22EE11 pKa = 4.25AQWNQRR17 pKa = 11.84STKK20 pKa = 9.94KK21 pKa = 10.25KK22 pKa = 10.28ASAPKK27 pKa = 9.94AKK29 pKa = 10.2KK30 pKa = 10.35AGGKK34 pKa = 8.85GEE36 pKa = 4.18KK37 pKa = 9.62PSLQIQTLLHH47 pKa = 6.55SGDD50 pKa = 3.49TMITVPSGGVCDD62 pKa = 6.17LINTYY67 pKa = 11.02ARR69 pKa = 11.84GSDD72 pKa = 3.04EE73 pKa = 4.39GNRR76 pKa = 11.84HH77 pKa = 4.54TSEE80 pKa = 3.79TLTYY84 pKa = 10.21KK85 pKa = 10.8VGVDD89 pKa = 3.04YY90 pKa = 11.43HH91 pKa = 6.46FVADD95 pKa = 4.09AASCKK100 pKa = 9.91YY101 pKa = 10.2SNRR104 pKa = 11.84GTGVMWLVYY113 pKa = 9.39DD114 pKa = 4.05TTPGGNAPTTKK125 pKa = 10.51DD126 pKa = 2.52IFAYY130 pKa = 10.59SDD132 pKa = 3.75ALKK135 pKa = 10.5AWPTTWKK142 pKa = 10.31VSRR145 pKa = 11.84EE146 pKa = 3.85LCHH149 pKa = 6.49RR150 pKa = 11.84FVVKK154 pKa = 10.41RR155 pKa = 11.84RR156 pKa = 11.84WLFTMEE162 pKa = 3.72TDD164 pKa = 3.84GRR166 pKa = 11.84IGSDD170 pKa = 3.46TPPSNASWPPCKK182 pKa = 10.18RR183 pKa = 11.84NVDD186 pKa = 4.1FHH188 pKa = 7.43KK189 pKa = 9.79FTSGLGVRR197 pKa = 11.84TQWKK201 pKa = 8.76NVTDD205 pKa = 4.02GGVGAIQRR213 pKa = 11.84GALYY217 pKa = 10.5LVIAPGNGITFTAHH231 pKa = 5.19GQTRR235 pKa = 11.84LYY237 pKa = 9.73FKK239 pKa = 10.92SVGNQQ244 pKa = 2.8
Molecular weight: 26.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
617 |
101 |
272 |
205.7 |
23.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.159 ± 1.294 | 1.459 ± 0.139 |
5.511 ± 0.833 | 4.538 ± 1.189 |
3.566 ± 0.298 | 6.969 ± 2.09 |
2.593 ± 0.234 | 5.186 ± 1.031 |
6.159 ± 1.541 | 7.942 ± 1.516 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.783 ± 0.29 | 4.052 ± 0.626 |
6.483 ± 1.37 | 4.214 ± 1.051 |
5.511 ± 0.688 | 8.914 ± 1.542 |
6.969 ± 1.945 | 6.159 ± 0.971 |
2.431 ± 0.363 | 3.404 ± 0.178 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
