
Rhizobium sp. BK376
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
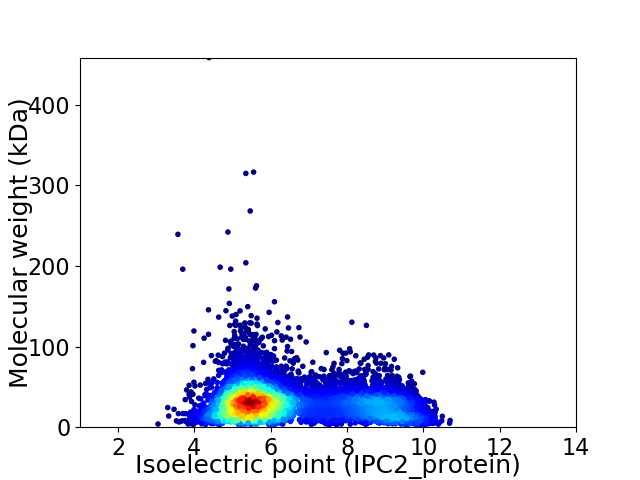
Virtual 2D-PAGE plot for 7471 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V2UB25|A0A4V2UB25_9RHIZ 4 5-dihydroxyphthalate decarboxylase OS=Rhizobium sp. BK376 OX=2512149 GN=EV561_10563 PE=4 SV=1
MM1 pKa = 8.19PIPQVSDD8 pKa = 3.86TIHH11 pKa = 6.87LNHH14 pKa = 6.88SDD16 pKa = 3.99AGDD19 pKa = 3.73GNAGNGGDD27 pKa = 4.48GYY29 pKa = 11.04NHH31 pKa = 7.13GDD33 pKa = 2.86ISYY36 pKa = 10.92NPIAFVANTQTVEE49 pKa = 3.76GAYY52 pKa = 9.52TDD54 pKa = 3.73LHH56 pKa = 7.48NGDD59 pKa = 5.12HH60 pKa = 5.9VWQAAGWGSGDD71 pKa = 3.45GGAGGASQAQNNGFFAAVTSGAGGAGGDD99 pKa = 3.73SHH101 pKa = 9.09SDD103 pKa = 3.23GDD105 pKa = 3.79QGSLSGGDD113 pKa = 3.47TAAVAAATTATQNTQLMADD132 pKa = 3.18QHH134 pKa = 5.39ATILAGVGGNGGNGNLAQGGDD155 pKa = 2.93ISAAVVHH162 pKa = 6.66SDD164 pKa = 4.32PEE166 pKa = 4.42TTTVNNALDD175 pKa = 3.65HH176 pKa = 5.89FVNAFGDD183 pKa = 3.4IDD185 pKa = 4.41LSHH188 pKa = 7.07LGSS191 pKa = 3.71
MM1 pKa = 8.19PIPQVSDD8 pKa = 3.86TIHH11 pKa = 6.87LNHH14 pKa = 6.88SDD16 pKa = 3.99AGDD19 pKa = 3.73GNAGNGGDD27 pKa = 4.48GYY29 pKa = 11.04NHH31 pKa = 7.13GDD33 pKa = 2.86ISYY36 pKa = 10.92NPIAFVANTQTVEE49 pKa = 3.76GAYY52 pKa = 9.52TDD54 pKa = 3.73LHH56 pKa = 7.48NGDD59 pKa = 5.12HH60 pKa = 5.9VWQAAGWGSGDD71 pKa = 3.45GGAGGASQAQNNGFFAAVTSGAGGAGGDD99 pKa = 3.73SHH101 pKa = 9.09SDD103 pKa = 3.23GDD105 pKa = 3.79QGSLSGGDD113 pKa = 3.47TAAVAAATTATQNTQLMADD132 pKa = 3.18QHH134 pKa = 5.39ATILAGVGGNGGNGNLAQGGDD155 pKa = 2.93ISAAVVHH162 pKa = 6.66SDD164 pKa = 4.32PEE166 pKa = 4.42TTTVNNALDD175 pKa = 3.65HH176 pKa = 5.89FVNAFGDD183 pKa = 3.4IDD185 pKa = 4.41LSHH188 pKa = 7.07LGSS191 pKa = 3.71
Molecular weight: 18.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V2U9Y4|A0A4V2U9Y4_9RHIZ Arsenite efflux membrane protein ArsB OS=Rhizobium sp. BK376 OX=2512149 GN=EV561_108112 PE=4 SV=1
MM1 pKa = 7.76LFRR4 pKa = 11.84EE5 pKa = 4.3PEE7 pKa = 3.64IPEE10 pKa = 4.14ASRR13 pKa = 11.84HH14 pKa = 5.31LPAGMEE20 pKa = 3.81ATGRR24 pKa = 11.84FSFDD28 pKa = 2.57EE29 pKa = 3.93SARR32 pKa = 11.84RR33 pKa = 11.84LNIEE37 pKa = 3.47ASVRR41 pKa = 11.84VLDD44 pKa = 4.33AVCGCGGRR52 pKa = 11.84LRR54 pKa = 11.84RR55 pKa = 11.84HH56 pKa = 5.78GRR58 pKa = 11.84RR59 pKa = 11.84QLVIADD65 pKa = 3.42VPYY68 pKa = 10.37RR69 pKa = 11.84GIPVRR74 pKa = 11.84IILVYY79 pKa = 9.83PRR81 pKa = 11.84LRR83 pKa = 11.84CGVCLTSRR91 pKa = 11.84RR92 pKa = 11.84PAIGDD97 pKa = 2.85IDD99 pKa = 3.59HH100 pKa = 7.03RR101 pKa = 11.84HH102 pKa = 4.96RR103 pKa = 11.84MSRR106 pKa = 11.84RR107 pKa = 11.84LLVHH111 pKa = 6.22LWEE114 pKa = 4.61EE115 pKa = 4.02ARR117 pKa = 11.84SRR119 pKa = 11.84PIADD123 pKa = 3.27IAHH126 pKa = 6.55SIGLDD131 pKa = 3.04EE132 pKa = 4.22KK133 pKa = 9.77TVRR136 pKa = 11.84NALASKK142 pKa = 10.26RR143 pKa = 11.84VSEE146 pKa = 4.36LDD148 pKa = 3.31NRR150 pKa = 11.84ICSVPQILGVARR162 pKa = 11.84CHH164 pKa = 5.53IKK166 pKa = 10.88GRR168 pKa = 11.84VTTVFWDD175 pKa = 3.26VGRR178 pKa = 11.84RR179 pKa = 11.84NLVDD183 pKa = 3.19IVFSDD188 pKa = 3.53RR189 pKa = 11.84ASDD192 pKa = 3.47VRR194 pKa = 11.84RR195 pKa = 11.84WLDD198 pKa = 3.27AFPEE202 pKa = 4.28RR203 pKa = 11.84EE204 pKa = 4.28LVSVVAIDD212 pKa = 3.5MVEE215 pKa = 4.01AYY217 pKa = 9.99HH218 pKa = 7.1RR219 pKa = 11.84AVQEE223 pKa = 4.03ALPEE227 pKa = 3.85ARR229 pKa = 11.84IAVHH233 pKa = 6.24PSGLRR238 pKa = 11.84KK239 pKa = 9.66RR240 pKa = 11.84FKK242 pKa = 10.73DD243 pKa = 3.44AADD246 pKa = 3.47KK247 pKa = 10.82VLKK250 pKa = 10.76NSTLTRR256 pKa = 11.84FKK258 pKa = 10.79ATDD261 pKa = 3.81PYY263 pKa = 10.43FAAHH267 pKa = 7.21RR268 pKa = 11.84ALCASGRR275 pKa = 11.84KK276 pKa = 8.45FQMQTLLDD284 pKa = 4.67LNPLMRR290 pKa = 11.84SAMEE294 pKa = 3.73ARR296 pKa = 11.84RR297 pKa = 11.84NYY299 pKa = 9.71HH300 pKa = 6.44DD301 pKa = 3.25VWKK304 pKa = 10.81ARR306 pKa = 11.84TPYY309 pKa = 10.22AAAIRR314 pKa = 11.84LEE316 pKa = 4.1SFLNNLSLLVLRR328 pKa = 11.84SFGDD332 pKa = 3.11IVSVVRR338 pKa = 11.84RR339 pKa = 11.84WARR342 pKa = 11.84QLADD346 pKa = 3.29STGIPGLKK354 pKa = 10.07GYY356 pKa = 9.92GAARR360 pKa = 11.84SMLDD364 pKa = 2.92RR365 pKa = 11.84AVANITAEE373 pKa = 4.12EE374 pKa = 4.43VEE376 pKa = 4.23NGSLGRR382 pKa = 11.84RR383 pKa = 11.84SPFDD387 pKa = 3.57DD388 pKa = 4.07CSIRR392 pKa = 11.84LCQCCDD398 pKa = 3.34HH399 pKa = 6.7EE400 pKa = 4.66NIRR403 pKa = 11.84YY404 pKa = 9.65GGGSRR409 pKa = 11.84LPRR412 pKa = 11.84SARR415 pKa = 11.84FGATILNPPVAKK427 pKa = 10.15RR428 pKa = 11.84FRR430 pKa = 11.84SIRR433 pKa = 11.84WICHH437 pKa = 4.23EE438 pKa = 5.05CNWSDD443 pKa = 4.33AEE445 pKa = 4.22VMLAQRR451 pKa = 11.84PNGLLRR457 pKa = 11.84ARR459 pKa = 4.41
MM1 pKa = 7.76LFRR4 pKa = 11.84EE5 pKa = 4.3PEE7 pKa = 3.64IPEE10 pKa = 4.14ASRR13 pKa = 11.84HH14 pKa = 5.31LPAGMEE20 pKa = 3.81ATGRR24 pKa = 11.84FSFDD28 pKa = 2.57EE29 pKa = 3.93SARR32 pKa = 11.84RR33 pKa = 11.84LNIEE37 pKa = 3.47ASVRR41 pKa = 11.84VLDD44 pKa = 4.33AVCGCGGRR52 pKa = 11.84LRR54 pKa = 11.84RR55 pKa = 11.84HH56 pKa = 5.78GRR58 pKa = 11.84RR59 pKa = 11.84QLVIADD65 pKa = 3.42VPYY68 pKa = 10.37RR69 pKa = 11.84GIPVRR74 pKa = 11.84IILVYY79 pKa = 9.83PRR81 pKa = 11.84LRR83 pKa = 11.84CGVCLTSRR91 pKa = 11.84RR92 pKa = 11.84PAIGDD97 pKa = 2.85IDD99 pKa = 3.59HH100 pKa = 7.03RR101 pKa = 11.84HH102 pKa = 4.96RR103 pKa = 11.84MSRR106 pKa = 11.84RR107 pKa = 11.84LLVHH111 pKa = 6.22LWEE114 pKa = 4.61EE115 pKa = 4.02ARR117 pKa = 11.84SRR119 pKa = 11.84PIADD123 pKa = 3.27IAHH126 pKa = 6.55SIGLDD131 pKa = 3.04EE132 pKa = 4.22KK133 pKa = 9.77TVRR136 pKa = 11.84NALASKK142 pKa = 10.26RR143 pKa = 11.84VSEE146 pKa = 4.36LDD148 pKa = 3.31NRR150 pKa = 11.84ICSVPQILGVARR162 pKa = 11.84CHH164 pKa = 5.53IKK166 pKa = 10.88GRR168 pKa = 11.84VTTVFWDD175 pKa = 3.26VGRR178 pKa = 11.84RR179 pKa = 11.84NLVDD183 pKa = 3.19IVFSDD188 pKa = 3.53RR189 pKa = 11.84ASDD192 pKa = 3.47VRR194 pKa = 11.84RR195 pKa = 11.84WLDD198 pKa = 3.27AFPEE202 pKa = 4.28RR203 pKa = 11.84EE204 pKa = 4.28LVSVVAIDD212 pKa = 3.5MVEE215 pKa = 4.01AYY217 pKa = 9.99HH218 pKa = 7.1RR219 pKa = 11.84AVQEE223 pKa = 4.03ALPEE227 pKa = 3.85ARR229 pKa = 11.84IAVHH233 pKa = 6.24PSGLRR238 pKa = 11.84KK239 pKa = 9.66RR240 pKa = 11.84FKK242 pKa = 10.73DD243 pKa = 3.44AADD246 pKa = 3.47KK247 pKa = 10.82VLKK250 pKa = 10.76NSTLTRR256 pKa = 11.84FKK258 pKa = 10.79ATDD261 pKa = 3.81PYY263 pKa = 10.43FAAHH267 pKa = 7.21RR268 pKa = 11.84ALCASGRR275 pKa = 11.84KK276 pKa = 8.45FQMQTLLDD284 pKa = 4.67LNPLMRR290 pKa = 11.84SAMEE294 pKa = 3.73ARR296 pKa = 11.84RR297 pKa = 11.84NYY299 pKa = 9.71HH300 pKa = 6.44DD301 pKa = 3.25VWKK304 pKa = 10.81ARR306 pKa = 11.84TPYY309 pKa = 10.22AAAIRR314 pKa = 11.84LEE316 pKa = 4.1SFLNNLSLLVLRR328 pKa = 11.84SFGDD332 pKa = 3.11IVSVVRR338 pKa = 11.84RR339 pKa = 11.84WARR342 pKa = 11.84QLADD346 pKa = 3.29STGIPGLKK354 pKa = 10.07GYY356 pKa = 9.92GAARR360 pKa = 11.84SMLDD364 pKa = 2.92RR365 pKa = 11.84AVANITAEE373 pKa = 4.12EE374 pKa = 4.43VEE376 pKa = 4.23NGSLGRR382 pKa = 11.84RR383 pKa = 11.84SPFDD387 pKa = 3.57DD388 pKa = 4.07CSIRR392 pKa = 11.84LCQCCDD398 pKa = 3.34HH399 pKa = 6.7EE400 pKa = 4.66NIRR403 pKa = 11.84YY404 pKa = 9.65GGGSRR409 pKa = 11.84LPRR412 pKa = 11.84SARR415 pKa = 11.84FGATILNPPVAKK427 pKa = 10.15RR428 pKa = 11.84FRR430 pKa = 11.84SIRR433 pKa = 11.84WICHH437 pKa = 4.23EE438 pKa = 5.05CNWSDD443 pKa = 4.33AEE445 pKa = 4.22VMLAQRR451 pKa = 11.84PNGLLRR457 pKa = 11.84ARR459 pKa = 4.41
Molecular weight: 51.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2212000 |
29 |
4713 |
296.1 |
32.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.556 ± 0.043 | 0.816 ± 0.008 |
5.764 ± 0.023 | 5.571 ± 0.03 |
3.926 ± 0.019 | 8.186 ± 0.028 |
2.07 ± 0.014 | 5.864 ± 0.018 |
3.768 ± 0.025 | 9.945 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.584 ± 0.012 | 2.94 ± 0.018 |
4.878 ± 0.022 | 3.189 ± 0.018 |
6.556 ± 0.032 | 6.094 ± 0.029 |
5.362 ± 0.023 | 7.286 ± 0.022 |
1.295 ± 0.011 | 2.351 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
