
Firmicutes bacterium CAG:194
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
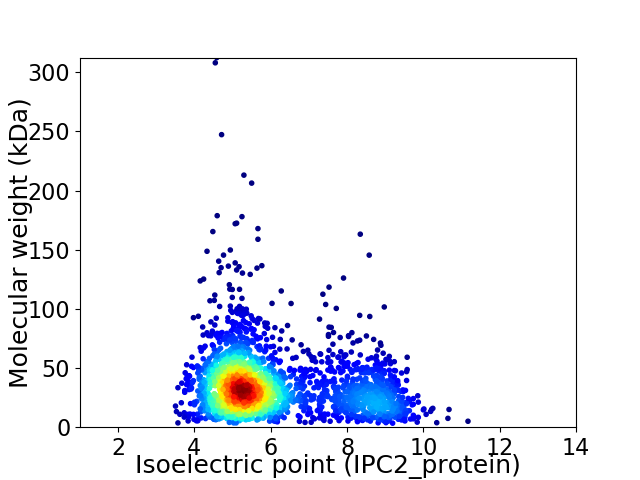
Virtual 2D-PAGE plot for 2310 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
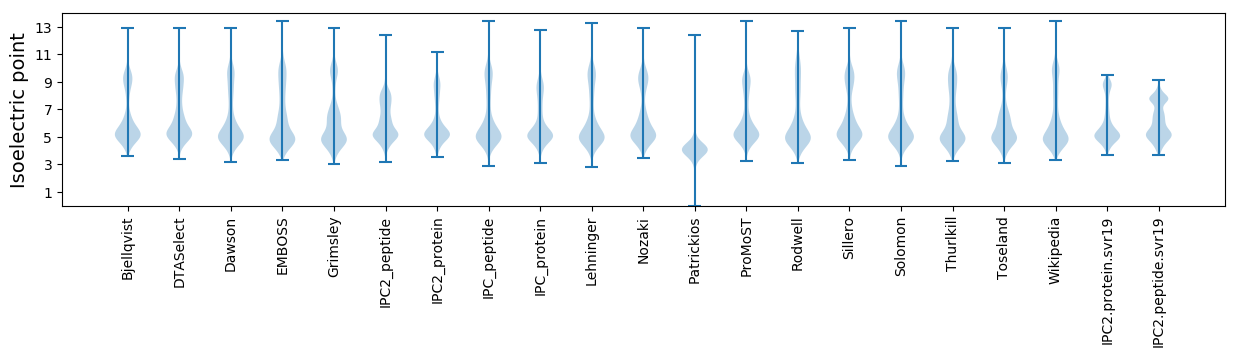
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5QP50|R5QP50_9FIRM Transcription-repair-coupling factor OS=Firmicutes bacterium CAG:194 OX=1263008 GN=mfd PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.46KK3 pKa = 10.37KK4 pKa = 10.38ILTLMLAACMVTALTACGSSKK25 pKa = 11.15EE26 pKa = 4.2EE27 pKa = 4.28GQSQDD32 pKa = 3.08TTEE35 pKa = 3.86VSGNGIISTKK45 pKa = 10.64DD46 pKa = 3.52YY47 pKa = 11.57DD48 pKa = 3.88PDD50 pKa = 5.49DD51 pKa = 3.88YY52 pKa = 12.03VKK54 pKa = 10.98LGDD57 pKa = 3.84YY58 pKa = 10.88EE59 pKa = 5.22GITVNTDD66 pKa = 2.21VYY68 pKa = 10.07TYY70 pKa = 11.0TDD72 pKa = 3.88ADD74 pKa = 3.03VDD76 pKa = 3.81AQFDD80 pKa = 3.96KK81 pKa = 11.04EE82 pKa = 3.72INYY85 pKa = 9.27YY86 pKa = 9.11VEE88 pKa = 5.09YY89 pKa = 9.88MDD91 pKa = 5.62AYY93 pKa = 10.82DD94 pKa = 3.63YY95 pKa = 9.46TATDD99 pKa = 3.38KK100 pKa = 10.48QTVEE104 pKa = 4.09TGDD107 pKa = 3.84TVNIDD112 pKa = 3.31YY113 pKa = 9.66TGTKK117 pKa = 10.22DD118 pKa = 3.96GVAFDD123 pKa = 4.3GGTAQGAHH131 pKa = 6.2LKK133 pKa = 10.05IGSGRR138 pKa = 11.84FIDD141 pKa = 4.31GFEE144 pKa = 4.89DD145 pKa = 3.92GLIGHH150 pKa = 7.45AVGEE154 pKa = 4.56TVNLDD159 pKa = 3.18LTFPEE164 pKa = 4.77NYY166 pKa = 10.29SNADD170 pKa = 3.43LAGAAVVFEE179 pKa = 4.27VKK181 pKa = 10.18INSIDD186 pKa = 3.6EE187 pKa = 4.53GKK189 pKa = 10.74LPEE192 pKa = 5.2INDD195 pKa = 3.82ALIASLNLGYY205 pKa = 9.07DD206 pKa = 3.19TVDD209 pKa = 2.79ACRR212 pKa = 11.84QNVEE216 pKa = 5.09DD217 pKa = 4.24YY218 pKa = 11.22LKK220 pKa = 10.79NACDD224 pKa = 4.2EE225 pKa = 4.48KK226 pKa = 11.51NSDD229 pKa = 3.65AKK231 pKa = 10.85KK232 pKa = 8.87SAVWNAVYY240 pKa = 8.24ATCEE244 pKa = 4.03VSDD247 pKa = 4.31APQEE251 pKa = 3.97LVDD254 pKa = 5.55DD255 pKa = 4.41IKK257 pKa = 10.57TRR259 pKa = 11.84IAANAQYY266 pKa = 10.59YY267 pKa = 9.37ADD269 pKa = 4.1YY270 pKa = 10.87YY271 pKa = 11.3KK272 pKa = 10.9VDD274 pKa = 3.92LATFITNYY282 pKa = 9.54MGLTEE287 pKa = 4.75EE288 pKa = 4.88EE289 pKa = 4.71YY290 pKa = 10.76EE291 pKa = 4.23KK292 pKa = 10.19QTAEE296 pKa = 4.13SAAEE300 pKa = 3.95SAKK303 pKa = 10.67EE304 pKa = 3.86KK305 pKa = 10.7LAIAAIAKK313 pKa = 9.62KK314 pKa = 10.51AGISLSDD321 pKa = 4.31DD322 pKa = 3.64DD323 pKa = 4.69VKK325 pKa = 11.22AAAEE329 pKa = 4.33TEE331 pKa = 4.21YY332 pKa = 11.33SDD334 pKa = 4.36YY335 pKa = 11.34GYY337 pKa = 11.14DD338 pKa = 3.78SADD341 pKa = 3.48ALLSAIGQGAYY352 pKa = 10.27YY353 pKa = 10.44DD354 pKa = 4.06YY355 pKa = 11.61VLSDD359 pKa = 3.62KK360 pKa = 11.14VNEE363 pKa = 3.97YY364 pKa = 10.95LIDD367 pKa = 3.43QVTIVEE373 pKa = 4.57NDD375 pKa = 4.22PISITEE381 pKa = 4.03TDD383 pKa = 3.21SAAADD388 pKa = 3.6TAEE391 pKa = 4.0EE392 pKa = 4.11DD393 pKa = 4.0VEE395 pKa = 4.91EE396 pKa = 4.14EE397 pKa = 4.1LQEE400 pKa = 4.18EE401 pKa = 4.68EE402 pKa = 4.61SVSGEE407 pKa = 3.97DD408 pKa = 4.65AAVTDD413 pKa = 3.7IAGTEE418 pKa = 3.97IEE420 pKa = 4.78AEE422 pKa = 4.19TEE424 pKa = 4.02EE425 pKa = 4.28
MM1 pKa = 7.44KK2 pKa = 10.46KK3 pKa = 10.37KK4 pKa = 10.38ILTLMLAACMVTALTACGSSKK25 pKa = 11.15EE26 pKa = 4.2EE27 pKa = 4.28GQSQDD32 pKa = 3.08TTEE35 pKa = 3.86VSGNGIISTKK45 pKa = 10.64DD46 pKa = 3.52YY47 pKa = 11.57DD48 pKa = 3.88PDD50 pKa = 5.49DD51 pKa = 3.88YY52 pKa = 12.03VKK54 pKa = 10.98LGDD57 pKa = 3.84YY58 pKa = 10.88EE59 pKa = 5.22GITVNTDD66 pKa = 2.21VYY68 pKa = 10.07TYY70 pKa = 11.0TDD72 pKa = 3.88ADD74 pKa = 3.03VDD76 pKa = 3.81AQFDD80 pKa = 3.96KK81 pKa = 11.04EE82 pKa = 3.72INYY85 pKa = 9.27YY86 pKa = 9.11VEE88 pKa = 5.09YY89 pKa = 9.88MDD91 pKa = 5.62AYY93 pKa = 10.82DD94 pKa = 3.63YY95 pKa = 9.46TATDD99 pKa = 3.38KK100 pKa = 10.48QTVEE104 pKa = 4.09TGDD107 pKa = 3.84TVNIDD112 pKa = 3.31YY113 pKa = 9.66TGTKK117 pKa = 10.22DD118 pKa = 3.96GVAFDD123 pKa = 4.3GGTAQGAHH131 pKa = 6.2LKK133 pKa = 10.05IGSGRR138 pKa = 11.84FIDD141 pKa = 4.31GFEE144 pKa = 4.89DD145 pKa = 3.92GLIGHH150 pKa = 7.45AVGEE154 pKa = 4.56TVNLDD159 pKa = 3.18LTFPEE164 pKa = 4.77NYY166 pKa = 10.29SNADD170 pKa = 3.43LAGAAVVFEE179 pKa = 4.27VKK181 pKa = 10.18INSIDD186 pKa = 3.6EE187 pKa = 4.53GKK189 pKa = 10.74LPEE192 pKa = 5.2INDD195 pKa = 3.82ALIASLNLGYY205 pKa = 9.07DD206 pKa = 3.19TVDD209 pKa = 2.79ACRR212 pKa = 11.84QNVEE216 pKa = 5.09DD217 pKa = 4.24YY218 pKa = 11.22LKK220 pKa = 10.79NACDD224 pKa = 4.2EE225 pKa = 4.48KK226 pKa = 11.51NSDD229 pKa = 3.65AKK231 pKa = 10.85KK232 pKa = 8.87SAVWNAVYY240 pKa = 8.24ATCEE244 pKa = 4.03VSDD247 pKa = 4.31APQEE251 pKa = 3.97LVDD254 pKa = 5.55DD255 pKa = 4.41IKK257 pKa = 10.57TRR259 pKa = 11.84IAANAQYY266 pKa = 10.59YY267 pKa = 9.37ADD269 pKa = 4.1YY270 pKa = 10.87YY271 pKa = 11.3KK272 pKa = 10.9VDD274 pKa = 3.92LATFITNYY282 pKa = 9.54MGLTEE287 pKa = 4.75EE288 pKa = 4.88EE289 pKa = 4.71YY290 pKa = 10.76EE291 pKa = 4.23KK292 pKa = 10.19QTAEE296 pKa = 4.13SAAEE300 pKa = 3.95SAKK303 pKa = 10.67EE304 pKa = 3.86KK305 pKa = 10.7LAIAAIAKK313 pKa = 9.62KK314 pKa = 10.51AGISLSDD321 pKa = 4.31DD322 pKa = 3.64DD323 pKa = 4.69VKK325 pKa = 11.22AAAEE329 pKa = 4.33TEE331 pKa = 4.21YY332 pKa = 11.33SDD334 pKa = 4.36YY335 pKa = 11.34GYY337 pKa = 11.14DD338 pKa = 3.78SADD341 pKa = 3.48ALLSAIGQGAYY352 pKa = 10.27YY353 pKa = 10.44DD354 pKa = 4.06YY355 pKa = 11.61VLSDD359 pKa = 3.62KK360 pKa = 11.14VNEE363 pKa = 3.97YY364 pKa = 10.95LIDD367 pKa = 3.43QVTIVEE373 pKa = 4.57NDD375 pKa = 4.22PISITEE381 pKa = 4.03TDD383 pKa = 3.21SAAADD388 pKa = 3.6TAEE391 pKa = 4.0EE392 pKa = 4.11DD393 pKa = 4.0VEE395 pKa = 4.91EE396 pKa = 4.14EE397 pKa = 4.1LQEE400 pKa = 4.18EE401 pKa = 4.68EE402 pKa = 4.61SVSGEE407 pKa = 3.97DD408 pKa = 4.65AAVTDD413 pKa = 3.7IAGTEE418 pKa = 3.97IEE420 pKa = 4.78AEE422 pKa = 4.19TEE424 pKa = 4.02EE425 pKa = 4.28
Molecular weight: 46.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5QPK8|R5QPK8_9FIRM 3-oxoacyl-[acyl-carrier-protein] synthase 2 OS=Firmicutes bacterium CAG:194 OX=1263008 GN=BN526_00745 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.81VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSAGGRR28 pKa = 11.84KK29 pKa = 8.81VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
763414 |
29 |
2771 |
330.5 |
37.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.931 ± 0.052 | 1.486 ± 0.019 |
5.764 ± 0.041 | 7.423 ± 0.066 |
3.947 ± 0.036 | 6.911 ± 0.047 |
1.719 ± 0.023 | 7.219 ± 0.052 |
7.007 ± 0.048 | 9.171 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.145 ± 0.028 | 4.073 ± 0.033 |
3.153 ± 0.028 | 3.694 ± 0.032 |
4.095 ± 0.036 | 5.592 ± 0.037 |
5.651 ± 0.046 | 6.916 ± 0.038 |
0.797 ± 0.015 | 4.306 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
