
Carnation Italian ringspot virus (CIRV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Tombusvirus
Average proteome isoelectric point is 8.09
Get precalculated fractions of proteins
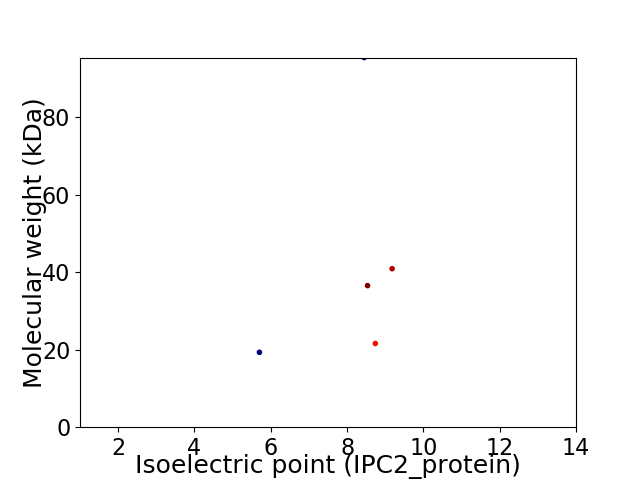
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
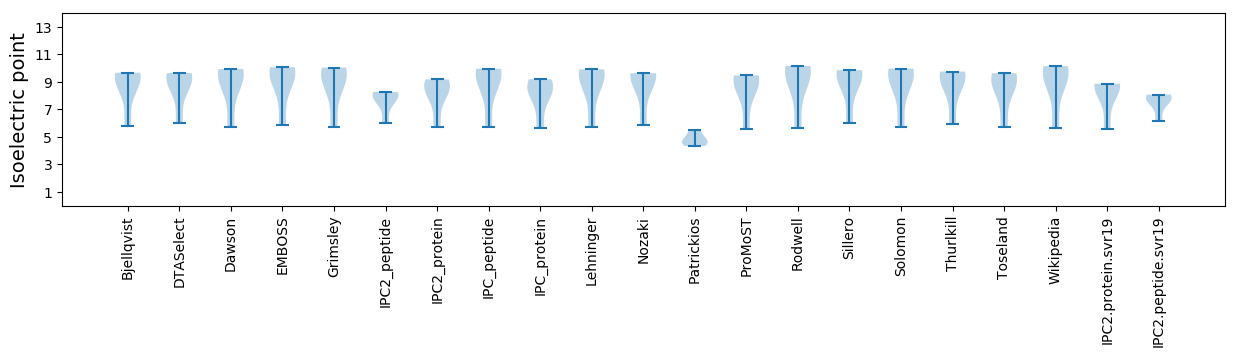
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q66104|P19_CIRV RNA silencing suppressor p19 OS=Carnation Italian ringspot virus OX=39443 GN=ORF4 PE=1 SV=1
MM1 pKa = 7.28EE2 pKa = 5.41RR3 pKa = 11.84AIQGNDD9 pKa = 2.71TRR11 pKa = 11.84EE12 pKa = 3.88QANGEE17 pKa = 4.11RR18 pKa = 11.84WDD20 pKa = 4.14GGSGGITSPFKK31 pKa = 11.15LPDD34 pKa = 3.7EE35 pKa = 4.69SPSWTEE41 pKa = 3.0WRR43 pKa = 11.84LYY45 pKa = 10.85NDD47 pKa = 3.84EE48 pKa = 4.46TNSNQDD54 pKa = 2.84NPLGFKK60 pKa = 10.19EE61 pKa = 3.92SWGFGKK67 pKa = 10.5VVFKK71 pKa = 10.8RR72 pKa = 11.84YY73 pKa = 9.61LRR75 pKa = 11.84YY76 pKa = 10.37DD77 pKa = 3.21RR78 pKa = 11.84TEE80 pKa = 3.87ASLHH84 pKa = 5.75RR85 pKa = 11.84VLGSWTGDD93 pKa = 3.38SVNYY97 pKa = 9.38AASRR101 pKa = 11.84FLGANQVGCTYY112 pKa = 10.56SIRR115 pKa = 11.84FRR117 pKa = 11.84GVSVTISGGSRR128 pKa = 11.84TLQHH132 pKa = 6.31LCEE135 pKa = 3.73MAIRR139 pKa = 11.84SKK141 pKa = 11.07QEE143 pKa = 3.79LLQLTPVEE151 pKa = 4.34VEE153 pKa = 4.42SNVSRR158 pKa = 11.84GCPEE162 pKa = 4.39GIEE165 pKa = 4.3TFKK168 pKa = 11.17KK169 pKa = 10.32EE170 pKa = 4.08SEE172 pKa = 4.1
MM1 pKa = 7.28EE2 pKa = 5.41RR3 pKa = 11.84AIQGNDD9 pKa = 2.71TRR11 pKa = 11.84EE12 pKa = 3.88QANGEE17 pKa = 4.11RR18 pKa = 11.84WDD20 pKa = 4.14GGSGGITSPFKK31 pKa = 11.15LPDD34 pKa = 3.7EE35 pKa = 4.69SPSWTEE41 pKa = 3.0WRR43 pKa = 11.84LYY45 pKa = 10.85NDD47 pKa = 3.84EE48 pKa = 4.46TNSNQDD54 pKa = 2.84NPLGFKK60 pKa = 10.19EE61 pKa = 3.92SWGFGKK67 pKa = 10.5VVFKK71 pKa = 10.8RR72 pKa = 11.84YY73 pKa = 9.61LRR75 pKa = 11.84YY76 pKa = 10.37DD77 pKa = 3.21RR78 pKa = 11.84TEE80 pKa = 3.87ASLHH84 pKa = 5.75RR85 pKa = 11.84VLGSWTGDD93 pKa = 3.38SVNYY97 pKa = 9.38AASRR101 pKa = 11.84FLGANQVGCTYY112 pKa = 10.56SIRR115 pKa = 11.84FRR117 pKa = 11.84GVSVTISGGSRR128 pKa = 11.84TLQHH132 pKa = 6.31LCEE135 pKa = 3.73MAIRR139 pKa = 11.84SKK141 pKa = 11.07QEE143 pKa = 3.79LLQLTPVEE151 pKa = 4.34VEE153 pKa = 4.42SNVSRR158 pKa = 11.84GCPEE162 pKa = 4.39GIEE165 pKa = 4.3TFKK168 pKa = 11.17KK169 pKa = 10.32EE170 pKa = 4.08SEE172 pKa = 4.1
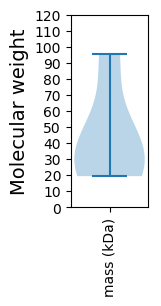
Molecular weight: 19.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q66103|Q66103_CIRV Movement protein OS=Carnation Italian ringspot virus OX=39443 GN=orf4 PE=3 SV=1
MM1 pKa = 7.25TMVLRR6 pKa = 11.84NSGTMVPQGGNKK18 pKa = 7.67MSKK21 pKa = 7.61MAKK24 pKa = 9.52NASQQGLQAGAAYY37 pKa = 10.82LMSNPKK43 pKa = 10.2GALKK47 pKa = 10.35KK48 pKa = 11.06GKK50 pKa = 9.6DD51 pKa = 2.9AWNYY55 pKa = 4.58MTAGGVPSGIVHH67 pKa = 6.14SGGMKK72 pKa = 9.33GAIMAPVAVTRR83 pKa = 11.84QLRR86 pKa = 11.84GSKK89 pKa = 9.96PKK91 pKa = 10.45FSGRR95 pKa = 11.84TSGSVTVSHH104 pKa = 7.15RR105 pKa = 11.84EE106 pKa = 4.4LITQVNNSTQFVVNGGVTGNLRR128 pKa = 11.84QVNPLNGTLFSWLPSIAANFDD149 pKa = 3.37QYY151 pKa = 11.33TFNSVTLHH159 pKa = 5.31YY160 pKa = 10.97VPLCATTEE168 pKa = 4.03TGRR171 pKa = 11.84VAMYY175 pKa = 10.13FDD177 pKa = 5.07KK178 pKa = 10.94DD179 pKa = 4.05SEE181 pKa = 4.34DD182 pKa = 4.11LEE184 pKa = 4.4PADD187 pKa = 4.12RR188 pKa = 11.84VEE190 pKa = 4.17LANYY194 pKa = 7.65ATLKK198 pKa = 8.78EE199 pKa = 4.05TAPWAEE205 pKa = 3.66AMLRR209 pKa = 11.84IPTDD213 pKa = 3.69RR214 pKa = 11.84IKK216 pKa = 10.68RR217 pKa = 11.84YY218 pKa = 10.24NDD220 pKa = 3.29DD221 pKa = 3.28SSVNDD226 pKa = 3.94RR227 pKa = 11.84KK228 pKa = 10.83LIDD231 pKa = 4.17LGQIGIATYY240 pKa = 10.34GGSGTNPVGDD250 pKa = 3.64VFISYY255 pKa = 10.53SVTLHH260 pKa = 6.42FPQPTAAGVQTRR272 pKa = 11.84RR273 pKa = 11.84LDD275 pKa = 3.56LTSVLDD281 pKa = 4.0TNVGPSYY288 pKa = 10.66TITSSTATVYY298 pKa = 9.64TVAFRR303 pKa = 11.84TVGTFMLFGAVRR315 pKa = 11.84STGSPVIGLSSANIAINSNTTLNTAGTAFSYY346 pKa = 7.1MTNVTVSALPASITFTVVGVITSSTSHH373 pKa = 6.65AVRR376 pKa = 11.84ASRR379 pKa = 11.84TNNADD384 pKa = 3.52MII386 pKa = 4.76
MM1 pKa = 7.25TMVLRR6 pKa = 11.84NSGTMVPQGGNKK18 pKa = 7.67MSKK21 pKa = 7.61MAKK24 pKa = 9.52NASQQGLQAGAAYY37 pKa = 10.82LMSNPKK43 pKa = 10.2GALKK47 pKa = 10.35KK48 pKa = 11.06GKK50 pKa = 9.6DD51 pKa = 2.9AWNYY55 pKa = 4.58MTAGGVPSGIVHH67 pKa = 6.14SGGMKK72 pKa = 9.33GAIMAPVAVTRR83 pKa = 11.84QLRR86 pKa = 11.84GSKK89 pKa = 9.96PKK91 pKa = 10.45FSGRR95 pKa = 11.84TSGSVTVSHH104 pKa = 7.15RR105 pKa = 11.84EE106 pKa = 4.4LITQVNNSTQFVVNGGVTGNLRR128 pKa = 11.84QVNPLNGTLFSWLPSIAANFDD149 pKa = 3.37QYY151 pKa = 11.33TFNSVTLHH159 pKa = 5.31YY160 pKa = 10.97VPLCATTEE168 pKa = 4.03TGRR171 pKa = 11.84VAMYY175 pKa = 10.13FDD177 pKa = 5.07KK178 pKa = 10.94DD179 pKa = 4.05SEE181 pKa = 4.34DD182 pKa = 4.11LEE184 pKa = 4.4PADD187 pKa = 4.12RR188 pKa = 11.84VEE190 pKa = 4.17LANYY194 pKa = 7.65ATLKK198 pKa = 8.78EE199 pKa = 4.05TAPWAEE205 pKa = 3.66AMLRR209 pKa = 11.84IPTDD213 pKa = 3.69RR214 pKa = 11.84IKK216 pKa = 10.68RR217 pKa = 11.84YY218 pKa = 10.24NDD220 pKa = 3.29DD221 pKa = 3.28SSVNDD226 pKa = 3.94RR227 pKa = 11.84KK228 pKa = 10.83LIDD231 pKa = 4.17LGQIGIATYY240 pKa = 10.34GGSGTNPVGDD250 pKa = 3.64VFISYY255 pKa = 10.53SVTLHH260 pKa = 6.42FPQPTAAGVQTRR272 pKa = 11.84RR273 pKa = 11.84LDD275 pKa = 3.56LTSVLDD281 pKa = 4.0TNVGPSYY288 pKa = 10.66TITSSTATVYY298 pKa = 9.64TVAFRR303 pKa = 11.84TVGTFMLFGAVRR315 pKa = 11.84STGSPVIGLSSANIAINSNTTLNTAGTAFSYY346 pKa = 7.1MTNVTVSALPASITFTVVGVITSSTSHH373 pKa = 6.65AVRR376 pKa = 11.84ASRR379 pKa = 11.84TNNADD384 pKa = 3.52MII386 pKa = 4.76
Molecular weight: 40.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1929 |
172 |
852 |
385.8 |
42.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.776 ± 1.007 | 1.555 ± 0.382 |
4.717 ± 0.286 | 5.547 ± 0.898 |
3.266 ± 0.266 | 7.932 ± 0.576 |
1.763 ± 0.414 | 4.303 ± 0.238 |
5.651 ± 0.67 | 8.917 ± 0.717 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.54 ± 0.277 | 4.355 ± 0.621 |
4.458 ± 0.173 | 2.696 ± 0.346 |
7.102 ± 0.63 | 6.843 ± 1.359 |
6.947 ± 1.18 | 8.605 ± 0.38 |
1.348 ± 0.217 | 3.629 ± 0.251 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
