
Chimpanzee associated porprismacovirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 7.8
Get precalculated fractions of proteins
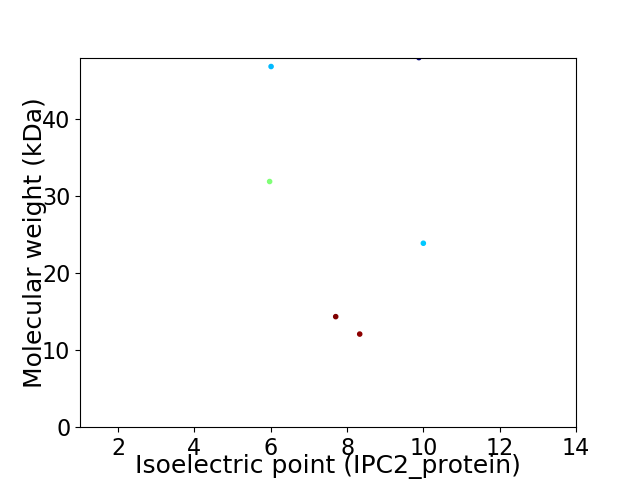
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
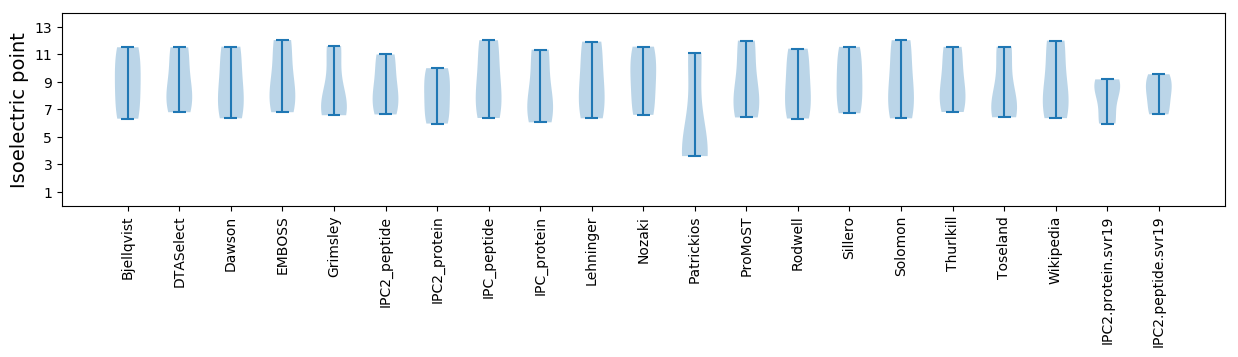
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D2WKE9|D2WKE9_9VIRU Putative replicase protein OS=Chimpanzee associated porprismacovirus 2 OX=2170110 GN=rep PE=4 SV=1
MM1 pKa = 7.45HH2 pKa = 7.8LFTEE6 pKa = 4.83VVSVDD11 pKa = 3.23IMTTMQRR18 pKa = 11.84THH20 pKa = 6.13TNAEE24 pKa = 4.22LIRR27 pKa = 11.84WFKK30 pKa = 10.53IFRR33 pKa = 11.84DD34 pKa = 3.64LDD36 pKa = 3.17IHH38 pKa = 6.15KK39 pKa = 9.68WVIGLEE45 pKa = 4.11EE46 pKa = 4.29GKK48 pKa = 10.53GGYY51 pKa = 7.67GHH53 pKa = 6.29WQVRR57 pKa = 11.84CNIRR61 pKa = 11.84VEE63 pKa = 4.38VVNDD67 pKa = 3.09WTAYY71 pKa = 9.53LRR73 pKa = 11.84AVFSWLGPISIWTEE87 pKa = 3.84EE88 pKa = 4.25CSDD91 pKa = 3.29KK92 pKa = 10.49YY93 pKa = 9.64TYY95 pKa = 7.33EE96 pKa = 4.28TKK98 pKa = 10.41EE99 pKa = 3.76GRR101 pKa = 11.84YY102 pKa = 6.08WASWDD107 pKa = 3.46TMGARR112 pKa = 11.84QQRR115 pKa = 11.84FGKK118 pKa = 9.08MRR120 pKa = 11.84WNQEE124 pKa = 3.52GAVQALQRR132 pKa = 11.84TNDD135 pKa = 3.34RR136 pKa = 11.84EE137 pKa = 4.07IVVWYY142 pKa = 9.93DD143 pKa = 3.11EE144 pKa = 4.06QGNMGKK150 pKa = 9.22SWLCGHH156 pKa = 7.23LFEE159 pKa = 5.62TGQAYY164 pKa = 8.57YY165 pKa = 9.68IPPYY169 pKa = 7.6MTSIQSMIQTVASLVLQDD187 pKa = 4.59RR188 pKa = 11.84DD189 pKa = 3.05SGYY192 pKa = 9.92PPRR195 pKa = 11.84PLIVIDD201 pKa = 5.23IPRR204 pKa = 11.84SWKK207 pKa = 9.63WSTEE211 pKa = 3.64LYY213 pKa = 9.64TAIEE217 pKa = 4.54AIKK220 pKa = 10.41DD221 pKa = 3.74GLIMDD226 pKa = 4.67PRR228 pKa = 11.84YY229 pKa = 9.71GARR232 pKa = 11.84PVNIHH237 pKa = 6.31GIKK240 pKa = 10.53VIVLTNTKK248 pKa = 9.98PKK250 pKa = 10.2LDD252 pKa = 3.97KK253 pKa = 10.88LSEE256 pKa = 4.3DD257 pKa = 2.82RR258 pKa = 11.84WVLYY262 pKa = 10.96DD263 pKa = 3.65PMDD266 pKa = 3.94YY267 pKa = 11.48PMMLL271 pKa = 3.63
MM1 pKa = 7.45HH2 pKa = 7.8LFTEE6 pKa = 4.83VVSVDD11 pKa = 3.23IMTTMQRR18 pKa = 11.84THH20 pKa = 6.13TNAEE24 pKa = 4.22LIRR27 pKa = 11.84WFKK30 pKa = 10.53IFRR33 pKa = 11.84DD34 pKa = 3.64LDD36 pKa = 3.17IHH38 pKa = 6.15KK39 pKa = 9.68WVIGLEE45 pKa = 4.11EE46 pKa = 4.29GKK48 pKa = 10.53GGYY51 pKa = 7.67GHH53 pKa = 6.29WQVRR57 pKa = 11.84CNIRR61 pKa = 11.84VEE63 pKa = 4.38VVNDD67 pKa = 3.09WTAYY71 pKa = 9.53LRR73 pKa = 11.84AVFSWLGPISIWTEE87 pKa = 3.84EE88 pKa = 4.25CSDD91 pKa = 3.29KK92 pKa = 10.49YY93 pKa = 9.64TYY95 pKa = 7.33EE96 pKa = 4.28TKK98 pKa = 10.41EE99 pKa = 3.76GRR101 pKa = 11.84YY102 pKa = 6.08WASWDD107 pKa = 3.46TMGARR112 pKa = 11.84QQRR115 pKa = 11.84FGKK118 pKa = 9.08MRR120 pKa = 11.84WNQEE124 pKa = 3.52GAVQALQRR132 pKa = 11.84TNDD135 pKa = 3.34RR136 pKa = 11.84EE137 pKa = 4.07IVVWYY142 pKa = 9.93DD143 pKa = 3.11EE144 pKa = 4.06QGNMGKK150 pKa = 9.22SWLCGHH156 pKa = 7.23LFEE159 pKa = 5.62TGQAYY164 pKa = 8.57YY165 pKa = 9.68IPPYY169 pKa = 7.6MTSIQSMIQTVASLVLQDD187 pKa = 4.59RR188 pKa = 11.84DD189 pKa = 3.05SGYY192 pKa = 9.92PPRR195 pKa = 11.84PLIVIDD201 pKa = 5.23IPRR204 pKa = 11.84SWKK207 pKa = 9.63WSTEE211 pKa = 3.64LYY213 pKa = 9.64TAIEE217 pKa = 4.54AIKK220 pKa = 10.41DD221 pKa = 3.74GLIMDD226 pKa = 4.67PRR228 pKa = 11.84YY229 pKa = 9.71GARR232 pKa = 11.84PVNIHH237 pKa = 6.31GIKK240 pKa = 10.53VIVLTNTKK248 pKa = 9.98PKK250 pKa = 10.2LDD252 pKa = 3.97KK253 pKa = 10.88LSEE256 pKa = 4.3DD257 pKa = 2.82RR258 pKa = 11.84WVLYY262 pKa = 10.96DD263 pKa = 3.65PMDD266 pKa = 3.94YY267 pKa = 11.48PMMLL271 pKa = 3.63
Molecular weight: 31.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D2WKE5|D2WKE5_9VIRU ORF3 OS=Chimpanzee associated porprismacovirus 2 OX=2170110 PE=4 SV=1
MM1 pKa = 7.83SIRR4 pKa = 11.84IKK6 pKa = 10.46IPQVLIKK13 pKa = 10.39GCPRR17 pKa = 11.84RR18 pKa = 11.84PLEE21 pKa = 3.62YY22 pKa = 10.61AFVYY26 pKa = 10.45GSGISGHH33 pKa = 6.23NDD35 pKa = 2.67NNAKK39 pKa = 10.21DD40 pKa = 3.62AYY42 pKa = 9.37QRR44 pKa = 11.84RR45 pKa = 11.84ANPLVQNLPRR55 pKa = 11.84SRR57 pKa = 11.84YY58 pKa = 7.66PQMGHH63 pKa = 6.47RR64 pKa = 11.84AGGRR68 pKa = 11.84QRR70 pKa = 11.84RR71 pKa = 11.84IRR73 pKa = 11.84TLAGQMQYY81 pKa = 10.62PGRR84 pKa = 11.84SGQRR88 pKa = 11.84LDD90 pKa = 4.67SIPKK94 pKa = 10.25GSILMARR101 pKa = 11.84TNLDD105 pKa = 3.03LDD107 pKa = 3.39RR108 pKa = 11.84GMFRR112 pKa = 11.84QVHH115 pKa = 5.86LRR117 pKa = 11.84DD118 pKa = 3.15QGRR121 pKa = 11.84QVLGIMGYY129 pKa = 10.48DD130 pKa = 3.52GSEE133 pKa = 3.87TTAVRR138 pKa = 11.84EE139 pKa = 4.08NEE141 pKa = 3.85MEE143 pKa = 4.01PRR145 pKa = 11.84RR146 pKa = 11.84GRR148 pKa = 11.84ASPTTDD154 pKa = 2.75EE155 pKa = 4.55RR156 pKa = 11.84PRR158 pKa = 11.84DD159 pKa = 3.59SGLVRR164 pKa = 11.84RR165 pKa = 11.84TRR167 pKa = 11.84QHH169 pKa = 4.86GQIVALRR176 pKa = 11.84APIRR180 pKa = 11.84DD181 pKa = 3.48RR182 pKa = 11.84AGLLHH187 pKa = 6.06TAVHH191 pKa = 6.94DD192 pKa = 3.6EE193 pKa = 4.27HH194 pKa = 6.69TKK196 pKa = 10.11HH197 pKa = 6.79DD198 pKa = 4.58SNRR201 pKa = 11.84SEE203 pKa = 4.19PSPTRR208 pKa = 11.84PRR210 pKa = 3.53
MM1 pKa = 7.83SIRR4 pKa = 11.84IKK6 pKa = 10.46IPQVLIKK13 pKa = 10.39GCPRR17 pKa = 11.84RR18 pKa = 11.84PLEE21 pKa = 3.62YY22 pKa = 10.61AFVYY26 pKa = 10.45GSGISGHH33 pKa = 6.23NDD35 pKa = 2.67NNAKK39 pKa = 10.21DD40 pKa = 3.62AYY42 pKa = 9.37QRR44 pKa = 11.84RR45 pKa = 11.84ANPLVQNLPRR55 pKa = 11.84SRR57 pKa = 11.84YY58 pKa = 7.66PQMGHH63 pKa = 6.47RR64 pKa = 11.84AGGRR68 pKa = 11.84QRR70 pKa = 11.84RR71 pKa = 11.84IRR73 pKa = 11.84TLAGQMQYY81 pKa = 10.62PGRR84 pKa = 11.84SGQRR88 pKa = 11.84LDD90 pKa = 4.67SIPKK94 pKa = 10.25GSILMARR101 pKa = 11.84TNLDD105 pKa = 3.03LDD107 pKa = 3.39RR108 pKa = 11.84GMFRR112 pKa = 11.84QVHH115 pKa = 5.86LRR117 pKa = 11.84DD118 pKa = 3.15QGRR121 pKa = 11.84QVLGIMGYY129 pKa = 10.48DD130 pKa = 3.52GSEE133 pKa = 3.87TTAVRR138 pKa = 11.84EE139 pKa = 4.08NEE141 pKa = 3.85MEE143 pKa = 4.01PRR145 pKa = 11.84RR146 pKa = 11.84GRR148 pKa = 11.84ASPTTDD154 pKa = 2.75EE155 pKa = 4.55RR156 pKa = 11.84PRR158 pKa = 11.84DD159 pKa = 3.59SGLVRR164 pKa = 11.84RR165 pKa = 11.84TRR167 pKa = 11.84QHH169 pKa = 4.86GQIVALRR176 pKa = 11.84APIRR180 pKa = 11.84DD181 pKa = 3.48RR182 pKa = 11.84AGLLHH187 pKa = 6.06TAVHH191 pKa = 6.94DD192 pKa = 3.6EE193 pKa = 4.27HH194 pKa = 6.69TKK196 pKa = 10.11HH197 pKa = 6.79DD198 pKa = 4.58SNRR201 pKa = 11.84SEE203 pKa = 4.19PSPTRR208 pKa = 11.84PRR210 pKa = 3.53
Molecular weight: 23.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1554 |
105 |
421 |
259.0 |
29.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.505 ± 0.821 | 1.351 ± 0.52 |
4.826 ± 0.502 | 4.054 ± 0.397 |
2.767 ± 0.408 | 6.435 ± 1.03 |
3.99 ± 0.583 | 7.722 ± 1.724 |
3.99 ± 0.332 | 8.43 ± 0.471 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.218 ± 0.556 | 4.762 ± 0.81 |
5.47 ± 0.375 | 4.054 ± 0.685 |
8.559 ± 1.881 | 8.366 ± 0.956 |
6.885 ± 0.395 | 5.856 ± 0.619 |
1.609 ± 0.789 | 3.153 ± 0.935 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
