
Pseudanabaena sp. PCC 7367
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Pseudanabaenaceae; Pseudanabaena; unclassified Pseudanabaena
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
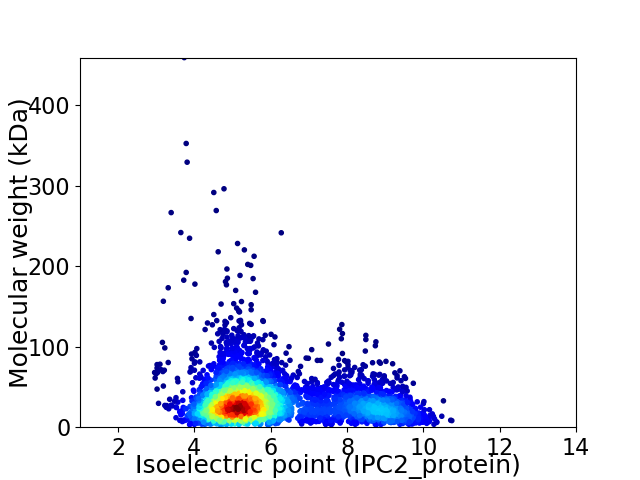
Virtual 2D-PAGE plot for 3824 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9SIH3|K9SIH3_9CYAN Putative cyanate ABC transporter substrate binding protein OS=Pseudanabaena sp. PCC 7367 OX=82654 GN=Pse7367_1717 PE=4 SV=1
MM1 pKa = 7.37SGSHH5 pKa = 6.33NPLEE9 pKa = 4.3AHH11 pKa = 7.28PSPTQAHH18 pKa = 6.49AFSPIPANTSVTNGSTSQVPNLAASQGAVDD48 pKa = 4.02FAFVQRR54 pKa = 11.84QGDD57 pKa = 3.51AYY59 pKa = 10.62YY60 pKa = 10.24PNNTGFFVGGDD71 pKa = 2.89IYY73 pKa = 11.11TPLVNDD79 pKa = 3.85VSTGGALGFLDD90 pKa = 5.87FYY92 pKa = 11.15VLPRR96 pKa = 11.84DD97 pKa = 3.53AVPQHH102 pKa = 4.94THH104 pKa = 4.2AHH106 pKa = 4.94EE107 pKa = 5.01AEE109 pKa = 3.91AKK111 pKa = 9.14YY112 pKa = 10.77VLEE115 pKa = 4.49GEE117 pKa = 4.74VNFDD121 pKa = 4.53LGPAQIRR128 pKa = 11.84VPTGSFIYY136 pKa = 10.48YY137 pKa = 9.8PIGRR141 pKa = 11.84QMGFTATDD149 pKa = 3.18QSARR153 pKa = 11.84ISVITTPGAPYY164 pKa = 10.76YY165 pKa = 9.81EE166 pKa = 4.38FAGVAVVDD174 pKa = 4.03EE175 pKa = 4.69QGNNTPPPQADD186 pKa = 2.84IGAIAAQLDD195 pKa = 4.08FGVVGQIIDD204 pKa = 3.88TYY206 pKa = 11.05GGGAFIPGVDD216 pKa = 3.39SAFQAGLQSPLLVVPDD232 pKa = 4.95LDD234 pKa = 6.2LIDD237 pKa = 4.43PAQLEE242 pKa = 4.31AVRR245 pKa = 11.84NVPGLNLQIFEE256 pKa = 3.96ISDD259 pKa = 3.24RR260 pKa = 11.84RR261 pKa = 11.84KK262 pKa = 9.68FDD264 pKa = 3.88GLFGTQNTSLIDD276 pKa = 3.8FEE278 pKa = 5.04EE279 pKa = 5.29SDD281 pKa = 4.01GNFAYY286 pKa = 10.63SQFNLARR293 pKa = 11.84DD294 pKa = 4.07DD295 pKa = 3.7RR296 pKa = 11.84HH297 pKa = 6.65SSNFLQAVINSDD309 pKa = 4.03QIVTTDD315 pKa = 3.28GVGVDD320 pKa = 3.86SGAFAYY326 pKa = 10.21ARR328 pKa = 11.84LHH330 pKa = 6.36AEE332 pKa = 3.85PNGTITYY339 pKa = 8.05EE340 pKa = 3.69ITINGLDD347 pKa = 3.2AGAYY351 pKa = 10.13LGDD354 pKa = 4.0GTPFTPDD361 pKa = 3.42PGDD364 pKa = 4.1DD365 pKa = 3.11LTAIHH370 pKa = 6.97LHH372 pKa = 4.67TAFRR376 pKa = 11.84GEE378 pKa = 4.27NGTHH382 pKa = 7.06AFNILGPGDD391 pKa = 4.31DD392 pKa = 4.89ADD394 pKa = 4.11LTVHH398 pKa = 6.43GNADD402 pKa = 3.47GSVTFSGVWDD412 pKa = 4.82DD413 pKa = 5.6SDD415 pKa = 3.85TTTDD419 pKa = 3.55TLPPPMSTKK428 pKa = 10.01PVSNFLPTLSNAAVGEE444 pKa = 4.12DD445 pKa = 2.78VGLYY449 pKa = 10.65VNIHH453 pKa = 5.6SNRR456 pKa = 11.84NSSGEE461 pKa = 3.48IRR463 pKa = 11.84GQVVGTTDD471 pKa = 3.98AFPASQVADD480 pKa = 3.35NHH482 pKa = 5.71LTFLVQEE489 pKa = 4.64GEE491 pKa = 4.28VAFQVNGNPFVAGVDD506 pKa = 3.5DD507 pKa = 4.23AVYY510 pKa = 10.16IAAGQNYY517 pKa = 10.22SIANLGNQNAVGLAAAVVDD536 pKa = 4.24SYY538 pKa = 11.68VPKK541 pKa = 10.33IGEE544 pKa = 4.29YY545 pKa = 10.28YY546 pKa = 10.58DD547 pKa = 4.07GNIDD551 pKa = 3.54FTASIRR557 pKa = 11.84TGDD560 pKa = 3.66EE561 pKa = 4.45FIQGNVVTTVPQAIGNGWIYY581 pKa = 9.81TWGAFGTDD589 pKa = 3.2DD590 pKa = 4.61FGNEE594 pKa = 3.88VPTSIGVTFTEE605 pKa = 4.96EE606 pKa = 3.68ALQDD610 pKa = 3.73AFVVDD615 pKa = 4.41DD616 pKa = 4.95PNNEE620 pKa = 4.02FPSSVPHH627 pKa = 6.48LAFPEE632 pKa = 3.89IFEE635 pKa = 4.12AARR638 pKa = 11.84VYY640 pKa = 11.05NILFPEE646 pKa = 4.87KK647 pKa = 9.92VQQSTPFNHH656 pKa = 6.22MGFYY660 pKa = 10.85ANSEE664 pKa = 3.81GHH666 pKa = 6.99APLDD670 pKa = 4.1IYY672 pKa = 10.93DD673 pKa = 4.46LPHH676 pKa = 6.7FDD678 pKa = 3.22VHH680 pKa = 6.72FFLDD684 pKa = 4.28TIEE687 pKa = 4.16EE688 pKa = 4.15RR689 pKa = 11.84DD690 pKa = 4.41LITGLPEE697 pKa = 5.02DD698 pKa = 3.99NANLFNLPEE707 pKa = 4.92PGFLPANYY715 pKa = 9.09IAPTIPGTDD724 pKa = 3.04IPATGDD730 pKa = 3.4ALQGIHH736 pKa = 5.39WVAGEE741 pKa = 4.01TPEE744 pKa = 4.49FNGEE748 pKa = 4.03VFDD751 pKa = 3.6QTFIFGSYY759 pKa = 10.27AGDD762 pKa = 3.53VNFWEE767 pKa = 4.27PMITRR772 pKa = 11.84DD773 pKa = 3.77FLEE776 pKa = 4.5SLSSARR782 pKa = 11.84QVITEE787 pKa = 3.9TTYY790 pKa = 11.27NIDD793 pKa = 2.98QPTRR797 pKa = 11.84FKK799 pKa = 11.0EE800 pKa = 3.81AGFYY804 pKa = 9.52PLEE807 pKa = 4.08YY808 pKa = 10.23GISYY812 pKa = 9.27NANHH816 pKa = 7.04GEE818 pKa = 4.21YY819 pKa = 10.0TVTLDD824 pKa = 3.28NFVIRR829 pKa = 11.84SADD832 pKa = 3.34GDD834 pKa = 3.78LSGFPGFDD842 pKa = 3.3GPPPFF847 pKa = 5.45
MM1 pKa = 7.37SGSHH5 pKa = 6.33NPLEE9 pKa = 4.3AHH11 pKa = 7.28PSPTQAHH18 pKa = 6.49AFSPIPANTSVTNGSTSQVPNLAASQGAVDD48 pKa = 4.02FAFVQRR54 pKa = 11.84QGDD57 pKa = 3.51AYY59 pKa = 10.62YY60 pKa = 10.24PNNTGFFVGGDD71 pKa = 2.89IYY73 pKa = 11.11TPLVNDD79 pKa = 3.85VSTGGALGFLDD90 pKa = 5.87FYY92 pKa = 11.15VLPRR96 pKa = 11.84DD97 pKa = 3.53AVPQHH102 pKa = 4.94THH104 pKa = 4.2AHH106 pKa = 4.94EE107 pKa = 5.01AEE109 pKa = 3.91AKK111 pKa = 9.14YY112 pKa = 10.77VLEE115 pKa = 4.49GEE117 pKa = 4.74VNFDD121 pKa = 4.53LGPAQIRR128 pKa = 11.84VPTGSFIYY136 pKa = 10.48YY137 pKa = 9.8PIGRR141 pKa = 11.84QMGFTATDD149 pKa = 3.18QSARR153 pKa = 11.84ISVITTPGAPYY164 pKa = 10.76YY165 pKa = 9.81EE166 pKa = 4.38FAGVAVVDD174 pKa = 4.03EE175 pKa = 4.69QGNNTPPPQADD186 pKa = 2.84IGAIAAQLDD195 pKa = 4.08FGVVGQIIDD204 pKa = 3.88TYY206 pKa = 11.05GGGAFIPGVDD216 pKa = 3.39SAFQAGLQSPLLVVPDD232 pKa = 4.95LDD234 pKa = 6.2LIDD237 pKa = 4.43PAQLEE242 pKa = 4.31AVRR245 pKa = 11.84NVPGLNLQIFEE256 pKa = 3.96ISDD259 pKa = 3.24RR260 pKa = 11.84RR261 pKa = 11.84KK262 pKa = 9.68FDD264 pKa = 3.88GLFGTQNTSLIDD276 pKa = 3.8FEE278 pKa = 5.04EE279 pKa = 5.29SDD281 pKa = 4.01GNFAYY286 pKa = 10.63SQFNLARR293 pKa = 11.84DD294 pKa = 4.07DD295 pKa = 3.7RR296 pKa = 11.84HH297 pKa = 6.65SSNFLQAVINSDD309 pKa = 4.03QIVTTDD315 pKa = 3.28GVGVDD320 pKa = 3.86SGAFAYY326 pKa = 10.21ARR328 pKa = 11.84LHH330 pKa = 6.36AEE332 pKa = 3.85PNGTITYY339 pKa = 8.05EE340 pKa = 3.69ITINGLDD347 pKa = 3.2AGAYY351 pKa = 10.13LGDD354 pKa = 4.0GTPFTPDD361 pKa = 3.42PGDD364 pKa = 4.1DD365 pKa = 3.11LTAIHH370 pKa = 6.97LHH372 pKa = 4.67TAFRR376 pKa = 11.84GEE378 pKa = 4.27NGTHH382 pKa = 7.06AFNILGPGDD391 pKa = 4.31DD392 pKa = 4.89ADD394 pKa = 4.11LTVHH398 pKa = 6.43GNADD402 pKa = 3.47GSVTFSGVWDD412 pKa = 4.82DD413 pKa = 5.6SDD415 pKa = 3.85TTTDD419 pKa = 3.55TLPPPMSTKK428 pKa = 10.01PVSNFLPTLSNAAVGEE444 pKa = 4.12DD445 pKa = 2.78VGLYY449 pKa = 10.65VNIHH453 pKa = 5.6SNRR456 pKa = 11.84NSSGEE461 pKa = 3.48IRR463 pKa = 11.84GQVVGTTDD471 pKa = 3.98AFPASQVADD480 pKa = 3.35NHH482 pKa = 5.71LTFLVQEE489 pKa = 4.64GEE491 pKa = 4.28VAFQVNGNPFVAGVDD506 pKa = 3.5DD507 pKa = 4.23AVYY510 pKa = 10.16IAAGQNYY517 pKa = 10.22SIANLGNQNAVGLAAAVVDD536 pKa = 4.24SYY538 pKa = 11.68VPKK541 pKa = 10.33IGEE544 pKa = 4.29YY545 pKa = 10.28YY546 pKa = 10.58DD547 pKa = 4.07GNIDD551 pKa = 3.54FTASIRR557 pKa = 11.84TGDD560 pKa = 3.66EE561 pKa = 4.45FIQGNVVTTVPQAIGNGWIYY581 pKa = 9.81TWGAFGTDD589 pKa = 3.2DD590 pKa = 4.61FGNEE594 pKa = 3.88VPTSIGVTFTEE605 pKa = 4.96EE606 pKa = 3.68ALQDD610 pKa = 3.73AFVVDD615 pKa = 4.41DD616 pKa = 4.95PNNEE620 pKa = 4.02FPSSVPHH627 pKa = 6.48LAFPEE632 pKa = 3.89IFEE635 pKa = 4.12AARR638 pKa = 11.84VYY640 pKa = 11.05NILFPEE646 pKa = 4.87KK647 pKa = 9.92VQQSTPFNHH656 pKa = 6.22MGFYY660 pKa = 10.85ANSEE664 pKa = 3.81GHH666 pKa = 6.99APLDD670 pKa = 4.1IYY672 pKa = 10.93DD673 pKa = 4.46LPHH676 pKa = 6.7FDD678 pKa = 3.22VHH680 pKa = 6.72FFLDD684 pKa = 4.28TIEE687 pKa = 4.16EE688 pKa = 4.15RR689 pKa = 11.84DD690 pKa = 4.41LITGLPEE697 pKa = 5.02DD698 pKa = 3.99NANLFNLPEE707 pKa = 4.92PGFLPANYY715 pKa = 9.09IAPTIPGTDD724 pKa = 3.04IPATGDD730 pKa = 3.4ALQGIHH736 pKa = 5.39WVAGEE741 pKa = 4.01TPEE744 pKa = 4.49FNGEE748 pKa = 4.03VFDD751 pKa = 3.6QTFIFGSYY759 pKa = 10.27AGDD762 pKa = 3.53VNFWEE767 pKa = 4.27PMITRR772 pKa = 11.84DD773 pKa = 3.77FLEE776 pKa = 4.5SLSSARR782 pKa = 11.84QVITEE787 pKa = 3.9TTYY790 pKa = 11.27NIDD793 pKa = 2.98QPTRR797 pKa = 11.84FKK799 pKa = 11.0EE800 pKa = 3.81AGFYY804 pKa = 9.52PLEE807 pKa = 4.08YY808 pKa = 10.23GISYY812 pKa = 9.27NANHH816 pKa = 7.04GEE818 pKa = 4.21YY819 pKa = 10.0TVTLDD824 pKa = 3.28NFVIRR829 pKa = 11.84SADD832 pKa = 3.34GDD834 pKa = 3.78LSGFPGFDD842 pKa = 3.3GPPPFF847 pKa = 5.45
Molecular weight: 90.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9SKN6|K9SKN6_9CYAN Uncharacterized protein OS=Pseudanabaena sp. PCC 7367 OX=82654 GN=Pse7367_2081 PE=4 SV=1
MM1 pKa = 7.54APKK4 pKa = 10.19DD5 pKa = 3.57YY6 pKa = 10.34RR7 pKa = 11.84RR8 pKa = 11.84CISCRR13 pKa = 11.84RR14 pKa = 11.84LDD16 pKa = 3.77HH17 pKa = 7.25RR18 pKa = 11.84DD19 pKa = 3.21HH20 pKa = 6.82FWRR23 pKa = 11.84VVRR26 pKa = 11.84SRR28 pKa = 11.84DD29 pKa = 3.24SSGQISVQLDD39 pKa = 3.08QGMGRR44 pKa = 11.84SAYY47 pKa = 8.73ICRR50 pKa = 11.84NADD53 pKa = 3.75CLKK56 pKa = 10.39QAQKK60 pKa = 10.82KK61 pKa = 9.0NRR63 pKa = 11.84LARR66 pKa = 11.84VLRR69 pKa = 11.84APVEE73 pKa = 3.74PTIYY77 pKa = 10.39EE78 pKa = 3.99RR79 pKa = 11.84LQARR83 pKa = 11.84LVAEE87 pKa = 4.01QAKK90 pKa = 10.38
MM1 pKa = 7.54APKK4 pKa = 10.19DD5 pKa = 3.57YY6 pKa = 10.34RR7 pKa = 11.84RR8 pKa = 11.84CISCRR13 pKa = 11.84RR14 pKa = 11.84LDD16 pKa = 3.77HH17 pKa = 7.25RR18 pKa = 11.84DD19 pKa = 3.21HH20 pKa = 6.82FWRR23 pKa = 11.84VVRR26 pKa = 11.84SRR28 pKa = 11.84DD29 pKa = 3.24SSGQISVQLDD39 pKa = 3.08QGMGRR44 pKa = 11.84SAYY47 pKa = 8.73ICRR50 pKa = 11.84NADD53 pKa = 3.75CLKK56 pKa = 10.39QAQKK60 pKa = 10.82KK61 pKa = 9.0NRR63 pKa = 11.84LARR66 pKa = 11.84VLRR69 pKa = 11.84APVEE73 pKa = 3.74PTIYY77 pKa = 10.39EE78 pKa = 3.99RR79 pKa = 11.84LQARR83 pKa = 11.84LVAEE87 pKa = 4.01QAKK90 pKa = 10.38
Molecular weight: 10.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1251467 |
29 |
4259 |
327.3 |
36.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.329 ± 0.036 | 0.919 ± 0.016 |
5.767 ± 0.043 | 6.007 ± 0.038 |
3.76 ± 0.026 | 6.697 ± 0.044 |
1.836 ± 0.021 | 6.758 ± 0.026 |
4.337 ± 0.038 | 10.514 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.032 ± 0.02 | 4.424 ± 0.035 |
4.771 ± 0.029 | 5.312 ± 0.038 |
5.171 ± 0.032 | 6.398 ± 0.032 |
5.372 ± 0.04 | 6.403 ± 0.032 |
1.305 ± 0.017 | 2.887 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
