
Ruminococcus sp. CAG:579
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Ruminococcus; environmental samples
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
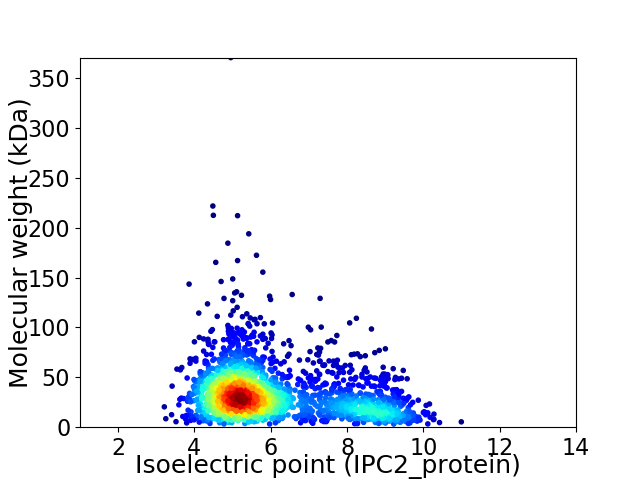
Virtual 2D-PAGE plot for 2155 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6CKK3|R6CKK3_9FIRM ACT domain protein OS=Ruminococcus sp. CAG:579 OX=1262963 GN=BN718_01721 PE=4 SV=1
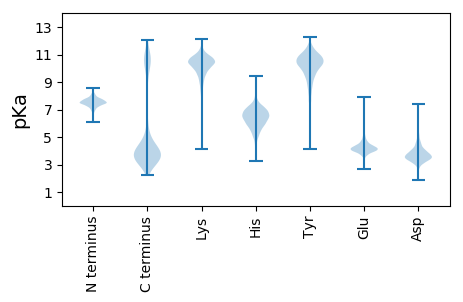
MM1 pKa = 8.02RR2 pKa = 11.84STKK5 pKa = 10.24SAALICAAVLVLMSMSACSKK25 pKa = 11.01SEE27 pKa = 3.86TAKK30 pKa = 10.24TNSSNADD37 pKa = 3.14SSSAKK42 pKa = 9.65STDD45 pKa = 2.97KK46 pKa = 11.32KK47 pKa = 10.84LDD49 pKa = 3.45VYY51 pKa = 10.87LSSDD55 pKa = 3.27GTPYY59 pKa = 11.15YY60 pKa = 10.66LADD63 pKa = 5.58DD64 pKa = 4.03GTKK67 pKa = 9.75MNLYY71 pKa = 10.43AYY73 pKa = 10.15ADD75 pKa = 4.03EE76 pKa = 6.13DD77 pKa = 4.6DD78 pKa = 4.87DD79 pKa = 4.18TAEE82 pKa = 4.96DD83 pKa = 3.87EE84 pKa = 4.89ASANYY89 pKa = 9.94IYY91 pKa = 10.83DD92 pKa = 3.74KK93 pKa = 11.35YY94 pKa = 11.34DD95 pKa = 3.7ADD97 pKa = 3.85GLKK100 pKa = 10.25FDD102 pKa = 5.11IPEE105 pKa = 3.75GWFADD110 pKa = 3.86TSFGSPMLLQQADD123 pKa = 3.63SDD125 pKa = 4.83DD126 pKa = 4.4EE127 pKa = 4.42INYY130 pKa = 10.03DD131 pKa = 3.54EE132 pKa = 5.51SIAIVDD138 pKa = 3.87TEE140 pKa = 4.45NVFDD144 pKa = 4.13SDD146 pKa = 3.4NGKK149 pKa = 8.21VTKK152 pKa = 9.34KK153 pKa = 8.3TVKK156 pKa = 10.32SYY158 pKa = 10.96FDD160 pKa = 3.56NYY162 pKa = 10.74LDD164 pKa = 3.36YY165 pKa = 11.17GYY167 pKa = 9.06YY168 pKa = 9.07TDD170 pKa = 4.46YY171 pKa = 10.83KK172 pKa = 10.36ISEE175 pKa = 4.32TGSMKK180 pKa = 10.42VSGVDD185 pKa = 3.04ANYY188 pKa = 10.56YY189 pKa = 10.71DD190 pKa = 4.54VITTISSALTNTDD203 pKa = 2.75SDD205 pKa = 3.67EE206 pKa = 4.33DD207 pKa = 4.12AEE209 pKa = 4.25PVVCKK214 pKa = 9.66TRR216 pKa = 11.84YY217 pKa = 9.75IITDD221 pKa = 3.69EE222 pKa = 4.27DD223 pKa = 4.06TSHH226 pKa = 7.48CMILSALNNDD236 pKa = 3.33EE237 pKa = 4.2SFGRR241 pKa = 11.84VVEE244 pKa = 4.93AYY246 pKa = 9.61EE247 pKa = 4.12KK248 pKa = 10.31MSGSLEE254 pKa = 4.18LPSS257 pKa = 3.81
MM1 pKa = 8.02RR2 pKa = 11.84STKK5 pKa = 10.24SAALICAAVLVLMSMSACSKK25 pKa = 11.01SEE27 pKa = 3.86TAKK30 pKa = 10.24TNSSNADD37 pKa = 3.14SSSAKK42 pKa = 9.65STDD45 pKa = 2.97KK46 pKa = 11.32KK47 pKa = 10.84LDD49 pKa = 3.45VYY51 pKa = 10.87LSSDD55 pKa = 3.27GTPYY59 pKa = 11.15YY60 pKa = 10.66LADD63 pKa = 5.58DD64 pKa = 4.03GTKK67 pKa = 9.75MNLYY71 pKa = 10.43AYY73 pKa = 10.15ADD75 pKa = 4.03EE76 pKa = 6.13DD77 pKa = 4.6DD78 pKa = 4.87DD79 pKa = 4.18TAEE82 pKa = 4.96DD83 pKa = 3.87EE84 pKa = 4.89ASANYY89 pKa = 9.94IYY91 pKa = 10.83DD92 pKa = 3.74KK93 pKa = 11.35YY94 pKa = 11.34DD95 pKa = 3.7ADD97 pKa = 3.85GLKK100 pKa = 10.25FDD102 pKa = 5.11IPEE105 pKa = 3.75GWFADD110 pKa = 3.86TSFGSPMLLQQADD123 pKa = 3.63SDD125 pKa = 4.83DD126 pKa = 4.4EE127 pKa = 4.42INYY130 pKa = 10.03DD131 pKa = 3.54EE132 pKa = 5.51SIAIVDD138 pKa = 3.87TEE140 pKa = 4.45NVFDD144 pKa = 4.13SDD146 pKa = 3.4NGKK149 pKa = 8.21VTKK152 pKa = 9.34KK153 pKa = 8.3TVKK156 pKa = 10.32SYY158 pKa = 10.96FDD160 pKa = 3.56NYY162 pKa = 10.74LDD164 pKa = 3.36YY165 pKa = 11.17GYY167 pKa = 9.06YY168 pKa = 9.07TDD170 pKa = 4.46YY171 pKa = 10.83KK172 pKa = 10.36ISEE175 pKa = 4.32TGSMKK180 pKa = 10.42VSGVDD185 pKa = 3.04ANYY188 pKa = 10.56YY189 pKa = 10.71DD190 pKa = 4.54VITTISSALTNTDD203 pKa = 2.75SDD205 pKa = 3.67EE206 pKa = 4.33DD207 pKa = 4.12AEE209 pKa = 4.25PVVCKK214 pKa = 9.66TRR216 pKa = 11.84YY217 pKa = 9.75IITDD221 pKa = 3.69EE222 pKa = 4.27DD223 pKa = 4.06TSHH226 pKa = 7.48CMILSALNNDD236 pKa = 3.33EE237 pKa = 4.2SFGRR241 pKa = 11.84VVEE244 pKa = 4.93AYY246 pKa = 9.61EE247 pKa = 4.12KK248 pKa = 10.31MSGSLEE254 pKa = 4.18LPSS257 pKa = 3.81
Molecular weight: 28.24 kDa
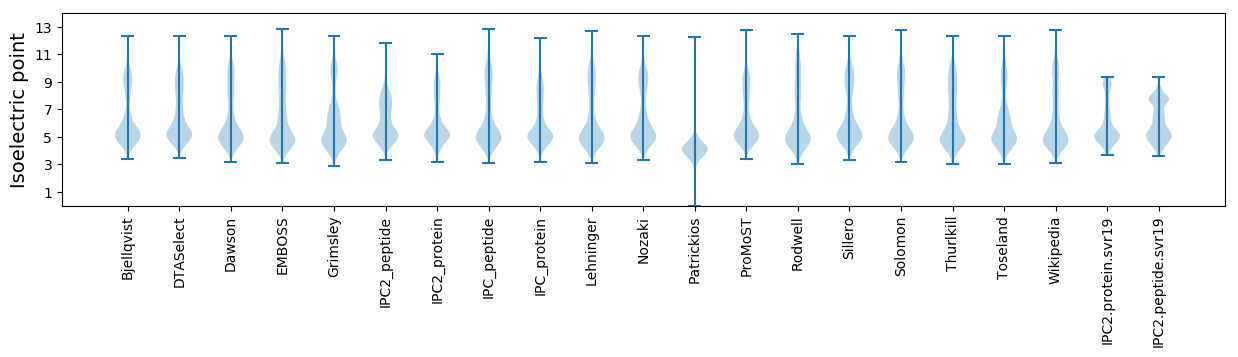
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6CNF1|R6CNF1_9FIRM Uncharacterized protein OS=Ruminococcus sp. CAG:579 OX=1262963 GN=BN718_00350 PE=4 SV=1
MM1 pKa = 7.67SNRR4 pKa = 11.84FCTGCGAPLTPDD16 pKa = 3.1MKK18 pKa = 10.47ICPRR22 pKa = 11.84CGKK25 pKa = 9.52VAAVSARR32 pKa = 11.84PRR34 pKa = 11.84PQQRR38 pKa = 11.84APRR41 pKa = 11.84PQQPVRR47 pKa = 11.84NVRR50 pKa = 11.84TVRR53 pKa = 11.84TGQTVQNGQTVRR65 pKa = 11.84NSQPQARR72 pKa = 11.84RR73 pKa = 11.84PQTAPQRR80 pKa = 11.84RR81 pKa = 11.84PAPMPRR87 pKa = 11.84PSPAPEE93 pKa = 3.87RR94 pKa = 11.84QEE96 pKa = 3.86RR97 pKa = 11.84APKK100 pKa = 10.0KK101 pKa = 8.81RR102 pKa = 11.84SKK104 pKa = 8.61KK105 pKa = 10.34ARR107 pKa = 11.84MIFHH111 pKa = 7.6AATAAVVLVGLYY123 pKa = 9.11FAIFFVQIFRR133 pKa = 11.84VKK135 pKa = 10.29ISTYY139 pKa = 10.06DD140 pKa = 3.21FKK142 pKa = 11.28TEE144 pKa = 3.66IKK146 pKa = 10.6LEE148 pKa = 4.08SSNYY152 pKa = 7.32GQAMEE157 pKa = 5.14SYY159 pKa = 9.88FEE161 pKa = 4.07SGKK164 pKa = 10.08WSVNPLTGTCTYY176 pKa = 10.63TGTTKK181 pKa = 10.6HH182 pKa = 5.91GDD184 pKa = 3.34EE185 pKa = 4.35YY186 pKa = 10.91EE187 pKa = 4.09IVYY190 pKa = 7.75TARR193 pKa = 11.84LKK195 pKa = 11.02VDD197 pKa = 3.46VKK199 pKa = 11.08NISINGKK206 pKa = 7.51DD207 pKa = 3.36VRR209 pKa = 11.84ADD211 pKa = 4.33RR212 pKa = 11.84IEE214 pKa = 4.07STLMGMFII222 pKa = 4.87
MM1 pKa = 7.67SNRR4 pKa = 11.84FCTGCGAPLTPDD16 pKa = 3.1MKK18 pKa = 10.47ICPRR22 pKa = 11.84CGKK25 pKa = 9.52VAAVSARR32 pKa = 11.84PRR34 pKa = 11.84PQQRR38 pKa = 11.84APRR41 pKa = 11.84PQQPVRR47 pKa = 11.84NVRR50 pKa = 11.84TVRR53 pKa = 11.84TGQTVQNGQTVRR65 pKa = 11.84NSQPQARR72 pKa = 11.84RR73 pKa = 11.84PQTAPQRR80 pKa = 11.84RR81 pKa = 11.84PAPMPRR87 pKa = 11.84PSPAPEE93 pKa = 3.87RR94 pKa = 11.84QEE96 pKa = 3.86RR97 pKa = 11.84APKK100 pKa = 10.0KK101 pKa = 8.81RR102 pKa = 11.84SKK104 pKa = 8.61KK105 pKa = 10.34ARR107 pKa = 11.84MIFHH111 pKa = 7.6AATAAVVLVGLYY123 pKa = 9.11FAIFFVQIFRR133 pKa = 11.84VKK135 pKa = 10.29ISTYY139 pKa = 10.06DD140 pKa = 3.21FKK142 pKa = 11.28TEE144 pKa = 3.66IKK146 pKa = 10.6LEE148 pKa = 4.08SSNYY152 pKa = 7.32GQAMEE157 pKa = 5.14SYY159 pKa = 9.88FEE161 pKa = 4.07SGKK164 pKa = 10.08WSVNPLTGTCTYY176 pKa = 10.63TGTTKK181 pKa = 10.6HH182 pKa = 5.91GDD184 pKa = 3.34EE185 pKa = 4.35YY186 pKa = 10.91EE187 pKa = 4.09IVYY190 pKa = 7.75TARR193 pKa = 11.84LKK195 pKa = 11.02VDD197 pKa = 3.46VKK199 pKa = 11.08NISINGKK206 pKa = 7.51DD207 pKa = 3.36VRR209 pKa = 11.84ADD211 pKa = 4.33RR212 pKa = 11.84IEE214 pKa = 4.07STLMGMFII222 pKa = 4.87
Molecular weight: 24.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
683710 |
29 |
3445 |
317.3 |
35.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.84 ± 0.063 | 1.718 ± 0.027 |
6.194 ± 0.046 | 7.238 ± 0.056 |
4.076 ± 0.037 | 7.112 ± 0.051 |
1.553 ± 0.022 | 6.954 ± 0.04 |
6.924 ± 0.048 | 8.245 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.898 ± 0.026 | 4.397 ± 0.045 |
3.312 ± 0.033 | 2.754 ± 0.026 |
4.369 ± 0.052 | 6.411 ± 0.053 |
5.559 ± 0.051 | 6.523 ± 0.045 |
0.803 ± 0.02 | 4.106 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
