
Epizootic hemorrhagic disease virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
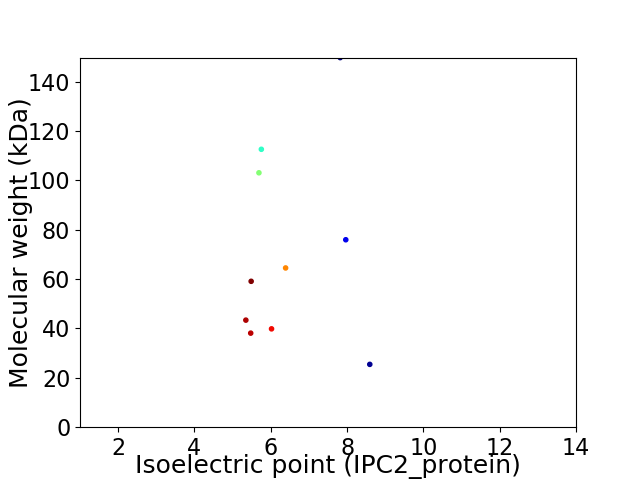
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
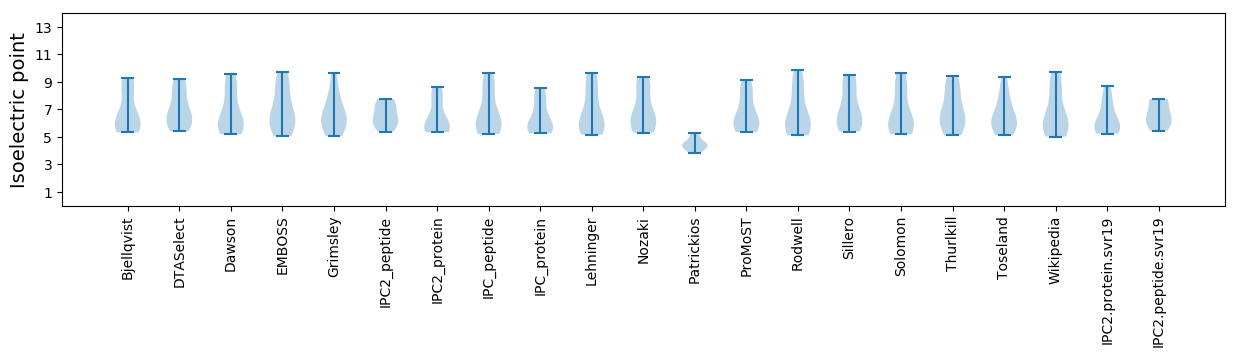
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V9SB81|V9SB81_9REOV Outer capsid protein VP2 OS=Epizootic hemorrhagic disease virus OX=40054 GN=VP2 PE=3 SV=1
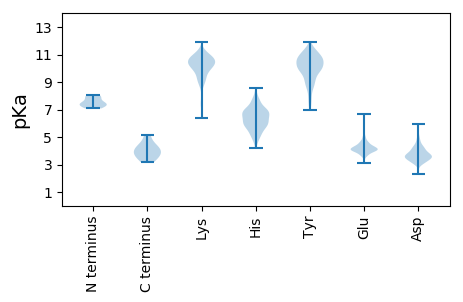
MM1 pKa = 7.45DD2 pKa = 5.55TIAARR7 pKa = 11.84ALTVIKK13 pKa = 10.52ACNTLKK19 pKa = 10.36EE20 pKa = 3.91VRR22 pKa = 11.84IVVEE26 pKa = 4.5SNVLEE31 pKa = 4.06ILGIAINRR39 pKa = 11.84YY40 pKa = 9.25NGLTLRR46 pKa = 11.84SVTMRR51 pKa = 11.84PTSQEE56 pKa = 3.34QRR58 pKa = 11.84NEE60 pKa = 3.99MFFMCLDD67 pKa = 3.66MVLAAANLNIGNISPDD83 pKa = 3.79YY84 pKa = 9.87IQNLATIGVLATPEE98 pKa = 3.68IPYY101 pKa = 10.22TMEE104 pKa = 3.9SANEE108 pKa = 3.85IARR111 pKa = 11.84MSGEE115 pKa = 4.11TGTWGPDD122 pKa = 3.13RR123 pKa = 11.84QPFGYY128 pKa = 9.89FLTAAEE134 pKa = 4.56VTQHH138 pKa = 4.89GRR140 pKa = 11.84FRR142 pKa = 11.84LRR144 pKa = 11.84AGQNITAAYY153 pKa = 9.63VSSTLAQVSMNAGARR168 pKa = 11.84GDD170 pKa = 3.65IQALFQNQNDD180 pKa = 4.44PIMIYY185 pKa = 9.16FVWRR189 pKa = 11.84RR190 pKa = 11.84IGTFSSAAGNAQDD203 pKa = 3.88TPQGVTLDD211 pKa = 3.59VGGVNMRR218 pKa = 11.84AGVIVAYY225 pKa = 10.33DD226 pKa = 3.42GQAPVNVNNPGAGPGMIEE244 pKa = 4.09IEE246 pKa = 4.28VIYY249 pKa = 10.36YY250 pKa = 10.66LSLDD254 pKa = 3.53KK255 pKa = 11.32TMTQYY260 pKa = 11.23PSLQAQIFNVYY271 pKa = 9.9SYY273 pKa = 11.5KK274 pKa = 10.76NPLWHH279 pKa = 6.91GLRR282 pKa = 11.84AAILNRR288 pKa = 11.84TTLPNNIPPIYY299 pKa = 9.64PPNDD303 pKa = 3.53RR304 pKa = 11.84EE305 pKa = 4.24NVLLLILLSALADD318 pKa = 3.8AFSVLAPDD326 pKa = 4.47FNLFGVVPIQGPINRR341 pKa = 11.84AVAQNAYY348 pKa = 7.85MM349 pKa = 4.54
MM1 pKa = 7.45DD2 pKa = 5.55TIAARR7 pKa = 11.84ALTVIKK13 pKa = 10.52ACNTLKK19 pKa = 10.36EE20 pKa = 3.91VRR22 pKa = 11.84IVVEE26 pKa = 4.5SNVLEE31 pKa = 4.06ILGIAINRR39 pKa = 11.84YY40 pKa = 9.25NGLTLRR46 pKa = 11.84SVTMRR51 pKa = 11.84PTSQEE56 pKa = 3.34QRR58 pKa = 11.84NEE60 pKa = 3.99MFFMCLDD67 pKa = 3.66MVLAAANLNIGNISPDD83 pKa = 3.79YY84 pKa = 9.87IQNLATIGVLATPEE98 pKa = 3.68IPYY101 pKa = 10.22TMEE104 pKa = 3.9SANEE108 pKa = 3.85IARR111 pKa = 11.84MSGEE115 pKa = 4.11TGTWGPDD122 pKa = 3.13RR123 pKa = 11.84QPFGYY128 pKa = 9.89FLTAAEE134 pKa = 4.56VTQHH138 pKa = 4.89GRR140 pKa = 11.84FRR142 pKa = 11.84LRR144 pKa = 11.84AGQNITAAYY153 pKa = 9.63VSSTLAQVSMNAGARR168 pKa = 11.84GDD170 pKa = 3.65IQALFQNQNDD180 pKa = 4.44PIMIYY185 pKa = 9.16FVWRR189 pKa = 11.84RR190 pKa = 11.84IGTFSSAAGNAQDD203 pKa = 3.88TPQGVTLDD211 pKa = 3.59VGGVNMRR218 pKa = 11.84AGVIVAYY225 pKa = 10.33DD226 pKa = 3.42GQAPVNVNNPGAGPGMIEE244 pKa = 4.09IEE246 pKa = 4.28VIYY249 pKa = 10.36YY250 pKa = 10.66LSLDD254 pKa = 3.53KK255 pKa = 11.32TMTQYY260 pKa = 11.23PSLQAQIFNVYY271 pKa = 9.9SYY273 pKa = 11.5KK274 pKa = 10.76NPLWHH279 pKa = 6.91GLRR282 pKa = 11.84AAILNRR288 pKa = 11.84TTLPNNIPPIYY299 pKa = 9.64PPNDD303 pKa = 3.53RR304 pKa = 11.84EE305 pKa = 4.24NVLLLILLSALADD318 pKa = 3.8AFSVLAPDD326 pKa = 4.47FNLFGVVPIQGPINRR341 pKa = 11.84AVAQNAYY348 pKa = 7.85MM349 pKa = 4.54
Molecular weight: 38.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V9SB52|V9SB52_9REOV Core protein VP7 OS=Epizootic hemorrhagic disease virus OX=40054 GN=VP7 PE=3 SV=1
MM1 pKa = 7.88LSRR4 pKa = 11.84LVPGVEE10 pKa = 4.01TRR12 pKa = 11.84IEE14 pKa = 4.02MKK16 pKa = 10.77ASDD19 pKa = 4.27EE20 pKa = 4.26VSLVPYY26 pKa = 9.71QEE28 pKa = 4.0QVRR31 pKa = 11.84PPSYY35 pKa = 10.68VPSAPIPTAMPKK47 pKa = 9.97VALDD51 pKa = 3.69ILDD54 pKa = 3.94KK55 pKa = 11.5AMSNQTGATMAQKK68 pKa = 10.38VEE70 pKa = 4.11KK71 pKa = 10.19VAYY74 pKa = 9.76ASYY77 pKa = 11.54AEE79 pKa = 4.37AFRR82 pKa = 11.84DD83 pKa = 3.96DD84 pKa = 3.69LRR86 pKa = 11.84LRR88 pKa = 11.84QIKK91 pKa = 9.96KK92 pKa = 9.63HH93 pKa = 5.46VNEE96 pKa = 3.91QVLPKK101 pKa = 9.93MRR103 pKa = 11.84VEE105 pKa = 4.13LTAMKK110 pKa = 10.09RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84AMAHH117 pKa = 6.14MILIIAAVVALITSASTLTSDD138 pKa = 4.25LGVILKK144 pKa = 10.67HH145 pKa = 5.3NTTTEE150 pKa = 4.75AIQTYY155 pKa = 10.06IKK157 pKa = 9.49PFCAAFGIINLAATMVMMFMAKK179 pKa = 9.51NEE181 pKa = 4.26KK182 pKa = 10.04IINQQIDD189 pKa = 3.54HH190 pKa = 5.65TRR192 pKa = 11.84KK193 pKa = 10.24EE194 pKa = 4.06IMKK197 pKa = 9.93KK198 pKa = 10.03DD199 pKa = 3.6AYY201 pKa = 10.75NEE203 pKa = 3.94AVRR206 pKa = 11.84MSVTEE211 pKa = 3.98FSGIPLDD218 pKa = 4.54GFDD221 pKa = 5.09IPPEE225 pKa = 4.01LTRR228 pKa = 5.13
MM1 pKa = 7.88LSRR4 pKa = 11.84LVPGVEE10 pKa = 4.01TRR12 pKa = 11.84IEE14 pKa = 4.02MKK16 pKa = 10.77ASDD19 pKa = 4.27EE20 pKa = 4.26VSLVPYY26 pKa = 9.71QEE28 pKa = 4.0QVRR31 pKa = 11.84PPSYY35 pKa = 10.68VPSAPIPTAMPKK47 pKa = 9.97VALDD51 pKa = 3.69ILDD54 pKa = 3.94KK55 pKa = 11.5AMSNQTGATMAQKK68 pKa = 10.38VEE70 pKa = 4.11KK71 pKa = 10.19VAYY74 pKa = 9.76ASYY77 pKa = 11.54AEE79 pKa = 4.37AFRR82 pKa = 11.84DD83 pKa = 3.96DD84 pKa = 3.69LRR86 pKa = 11.84LRR88 pKa = 11.84QIKK91 pKa = 9.96KK92 pKa = 9.63HH93 pKa = 5.46VNEE96 pKa = 3.91QVLPKK101 pKa = 9.93MRR103 pKa = 11.84VEE105 pKa = 4.13LTAMKK110 pKa = 10.09RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84AMAHH117 pKa = 6.14MILIIAAVVALITSASTLTSDD138 pKa = 4.25LGVILKK144 pKa = 10.67HH145 pKa = 5.3NTTTEE150 pKa = 4.75AIQTYY155 pKa = 10.06IKK157 pKa = 9.49PFCAAFGIINLAATMVMMFMAKK179 pKa = 9.51NEE181 pKa = 4.26KK182 pKa = 10.04IINQQIDD189 pKa = 3.54HH190 pKa = 5.65TRR192 pKa = 11.84KK193 pKa = 10.24EE194 pKa = 4.06IMKK197 pKa = 9.93KK198 pKa = 10.03DD199 pKa = 3.6AYY201 pKa = 10.75NEE203 pKa = 3.94AVRR206 pKa = 11.84MSVTEE211 pKa = 3.98FSGIPLDD218 pKa = 4.54GFDD221 pKa = 5.09IPPEE225 pKa = 4.01LTRR228 pKa = 5.13
Molecular weight: 25.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6205 |
228 |
1302 |
620.5 |
71.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.591 ± 0.553 | 1.08 ± 0.226 |
5.882 ± 0.274 | 7.961 ± 0.511 |
3.884 ± 0.25 | 5.915 ± 0.34 |
2.047 ± 0.275 | 7.462 ± 0.401 |
6.269 ± 0.619 | 8.042 ± 0.359 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.626 ± 0.376 | 3.916 ± 0.308 |
3.981 ± 0.295 | 4.11 ± 0.32 |
7.107 ± 0.261 | 5.222 ± 0.377 |
4.996 ± 0.348 | 6.833 ± 0.225 |
1.193 ± 0.235 | 3.884 ± 0.249 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
