
Iris yellow spot virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Tospoviridae; Orthotospovirus; Iris yellow spot orthotospovirus
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
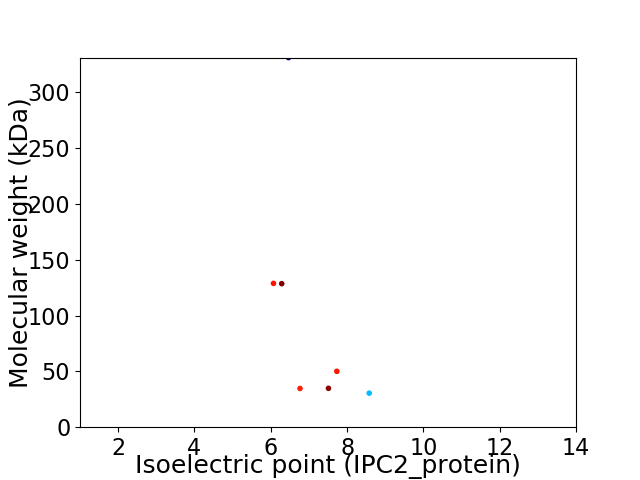
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0KQW8|C0KQW8_9VIRU Replicase OS=Iris yellow spot virus OX=60456 PE=4 SV=1
MM1 pKa = 7.27NLHH4 pKa = 6.51YY5 pKa = 10.95LLLYY9 pKa = 10.52CFGVYY14 pKa = 10.61LMFTITDD21 pKa = 3.57VLLMNNAEE29 pKa = 4.02DD30 pKa = 4.13HH31 pKa = 6.86EE32 pKa = 4.85DD33 pKa = 2.9IKK35 pKa = 11.1MEE37 pKa = 4.6LKK39 pKa = 10.43RR40 pKa = 11.84LKK42 pKa = 10.42DD43 pKa = 3.27RR44 pKa = 11.84YY45 pKa = 10.87KK46 pKa = 11.03NDD48 pKa = 3.56DD49 pKa = 3.93PEE51 pKa = 5.33DD52 pKa = 4.49LIEE55 pKa = 4.92NKK57 pKa = 10.16VIDD60 pKa = 4.4PGYY63 pKa = 10.13SFPSLDD69 pKa = 3.3PKK71 pKa = 10.53IPEE74 pKa = 4.67RR75 pKa = 11.84IPRR78 pKa = 11.84EE79 pKa = 3.71ATTNVPSTTNVPSTTDD95 pKa = 3.21RR96 pKa = 11.84QVVEE100 pKa = 4.6TLSCDD105 pKa = 3.9DD106 pKa = 4.15FDD108 pKa = 5.4KK109 pKa = 11.17SKK111 pKa = 11.07CIIKK115 pKa = 8.88GTSDD119 pKa = 3.71FNAHH123 pKa = 5.15YY124 pKa = 9.49QVKK127 pKa = 10.5GSDD130 pKa = 3.54SVIACITDD138 pKa = 3.33SEE140 pKa = 4.98KK141 pKa = 10.82IFDD144 pKa = 3.96ACLFDD149 pKa = 5.57SDD151 pKa = 3.42IKK153 pKa = 10.95RR154 pKa = 11.84RR155 pKa = 11.84TFLKK159 pKa = 10.43VPVVPVLKK167 pKa = 10.8LEE169 pKa = 4.2NKK171 pKa = 9.7RR172 pKa = 11.84VLEE175 pKa = 4.18VGSKK179 pKa = 10.11FFFVDD184 pKa = 3.69SSNDD188 pKa = 4.01PILSDD193 pKa = 4.32PKK195 pKa = 10.92SGMANPTVSRR205 pKa = 11.84LSVRR209 pKa = 11.84LSGDD213 pKa = 3.57CIINQVSMSSPYY225 pKa = 9.23MIKK228 pKa = 10.05IKK230 pKa = 9.66STEE233 pKa = 4.17TIGFAVKK240 pKa = 10.41NIKK243 pKa = 10.03NPQAINFQTASGDD256 pKa = 3.08KK257 pKa = 10.33SVYY260 pKa = 10.38FKK262 pKa = 10.52IDD264 pKa = 3.52EE265 pKa = 4.43LDD267 pKa = 3.69GNHH270 pKa = 6.31FFLCGDD276 pKa = 3.18KK277 pKa = 11.08SSFIKK282 pKa = 10.69KK283 pKa = 9.47VDD285 pKa = 3.46VPVRR289 pKa = 11.84NCVSKK294 pKa = 11.27YY295 pKa = 10.19ADD297 pKa = 3.58EE298 pKa = 4.34PKK300 pKa = 10.64KK301 pKa = 10.8IFFCTNFSYY310 pKa = 10.04FKK312 pKa = 10.28WFFTFLVITFPITWILWKK330 pKa = 10.46AKK332 pKa = 9.43NAFSIWYY339 pKa = 8.8DD340 pKa = 2.83IMGILTYY347 pKa = 10.28PILISINYY355 pKa = 8.04LWLYY359 pKa = 10.63FPFKK363 pKa = 10.63CKK365 pKa = 10.49SCGNLSFLSHH375 pKa = 6.22EE376 pKa = 4.38CSRR379 pKa = 11.84LCVCNQSKK387 pKa = 10.24PSRR390 pKa = 11.84EE391 pKa = 3.97HH392 pKa = 7.2SKK394 pKa = 10.57EE395 pKa = 3.73CYY397 pKa = 9.77LFHH400 pKa = 7.89KK401 pKa = 8.97DD402 pKa = 3.14TKK404 pKa = 9.93KK405 pKa = 9.73WRR407 pKa = 11.84SLSLIDD413 pKa = 3.64HH414 pKa = 6.51FQFIVNTKK422 pKa = 9.2ISSEE426 pKa = 3.9FLVFLTKK433 pKa = 9.98MLIAGVLMSCIPSSLALSSEE453 pKa = 4.12RR454 pKa = 11.84NVCVEE459 pKa = 3.55KK460 pKa = 10.91CFYY463 pKa = 11.22SKK465 pKa = 10.9DD466 pKa = 3.79LEE468 pKa = 4.38KK469 pKa = 10.66LTTDD473 pKa = 3.27KK474 pKa = 11.33DD475 pKa = 3.85GSIKK479 pKa = 10.72DD480 pKa = 3.99SIDD483 pKa = 3.2TCEE486 pKa = 4.11CSIGEE491 pKa = 4.81LITEE495 pKa = 4.4TVYY498 pKa = 10.87RR499 pKa = 11.84GGAPISRR506 pKa = 11.84AAIKK510 pKa = 10.14NDD512 pKa = 3.37CVSLSSKK519 pKa = 10.4CLNSSNQAEE528 pKa = 4.21NLFACRR534 pKa = 11.84HH535 pKa = 5.08GCNALSTLNKK545 pKa = 9.16IPKK548 pKa = 9.04VMYY551 pKa = 9.64DD552 pKa = 2.87RR553 pKa = 11.84GYY555 pKa = 11.02KK556 pKa = 10.05GVEE559 pKa = 3.7FSGNLTVLKK568 pKa = 10.16IANRR572 pKa = 11.84LRR574 pKa = 11.84KK575 pKa = 10.13GFVDD579 pKa = 3.45NQSEE583 pKa = 4.35VRR585 pKa = 11.84NLEE588 pKa = 4.19SKK590 pKa = 10.24VSKK593 pKa = 9.78EE594 pKa = 3.46LSYY597 pKa = 11.0FKK599 pKa = 10.56SLKK602 pKa = 10.27VDD604 pKa = 3.77DD605 pKa = 4.73VPPEE609 pKa = 3.92NLMPRR614 pKa = 11.84QSLVFSTEE622 pKa = 3.11VDD624 pKa = 3.05GKK626 pKa = 8.53YY627 pKa = 10.23RR628 pKa = 11.84YY629 pKa = 9.01MIEE632 pKa = 3.79MDD634 pKa = 3.16IKK636 pKa = 10.92KK637 pKa = 8.53EE638 pKa = 4.09TGSVFLLNDD647 pKa = 4.14DD648 pKa = 4.19SSHH651 pKa = 7.66IPMEE655 pKa = 3.59FMVYY659 pKa = 8.85VKK661 pKa = 10.53SVGVEE666 pKa = 3.2YY667 pKa = 10.4DD668 pKa = 2.72IRR670 pKa = 11.84YY671 pKa = 9.5KK672 pKa = 11.09YY673 pKa = 9.68STAKK677 pKa = 10.37VDD679 pKa = 3.74TTVTDD684 pKa = 3.76YY685 pKa = 11.68LSTCTGDD692 pKa = 4.76CKK694 pKa = 10.83DD695 pKa = 3.8CRR697 pKa = 11.84KK698 pKa = 10.47SKK700 pKa = 9.71TPSGHH705 pKa = 6.71NDD707 pKa = 2.85FCIQPTSWWGCEE719 pKa = 3.82EE720 pKa = 4.53LGCLAINEE728 pKa = 4.33GAICGHH734 pKa = 5.88CTNVFDD740 pKa = 6.71LSTLVNVYY748 pKa = 9.59QVVEE752 pKa = 3.77SHH754 pKa = 7.14IIAEE758 pKa = 4.15ICIKK762 pKa = 10.55SLNGYY767 pKa = 8.27DD768 pKa = 3.85CRR770 pKa = 11.84KK771 pKa = 10.01HH772 pKa = 6.79SDD774 pKa = 3.3RR775 pKa = 11.84APIQTDD781 pKa = 4.26YY782 pKa = 10.88YY783 pKa = 9.83QLDD786 pKa = 3.82MTVDD790 pKa = 3.42LHH792 pKa = 7.59NDD794 pKa = 3.21FMSTDD799 pKa = 3.18KK800 pKa = 11.32LFAVTKK806 pKa = 8.66NQKK809 pKa = 9.5ILTGSISDD817 pKa = 4.19LSDD820 pKa = 3.93FSSGAFGHH828 pKa = 5.83PQITVDD834 pKa = 3.79GTPLAVKK841 pKa = 9.2ATISRR846 pKa = 11.84DD847 pKa = 3.22QFTWSCSAVGSKK859 pKa = 10.0SVNIKK864 pKa = 9.92QCGLYY869 pKa = 9.56SYY871 pKa = 11.67NMIYY875 pKa = 10.99ALTQSKK881 pKa = 10.43DD882 pKa = 3.32FSVIDD887 pKa = 3.61EE888 pKa = 4.73EE889 pKa = 4.48NNKK892 pKa = 10.63LYY894 pKa = 9.93MIKK897 pKa = 10.51DD898 pKa = 3.6SLVGKK903 pKa = 9.61LKK905 pKa = 10.92VVIDD909 pKa = 3.75MPKK912 pKa = 10.71EE913 pKa = 4.01MFKK916 pKa = 9.53KK917 pKa = 9.2TPVKK921 pKa = 10.31PILSEE926 pKa = 4.02TKK928 pKa = 9.89IICSGCTKK936 pKa = 10.28CAMGLEE942 pKa = 4.51CVMEE946 pKa = 4.2YY947 pKa = 10.46TSDD950 pKa = 3.36TTFSSRR956 pKa = 11.84LKK958 pKa = 10.11MDD960 pKa = 3.74GCSFKK965 pKa = 10.77SDD967 pKa = 3.6QIGSYY972 pKa = 9.9IGPNKK977 pKa = 10.2KK978 pKa = 10.07LIKK981 pKa = 9.74AYY983 pKa = 10.36CSEE986 pKa = 4.21SVKK989 pKa = 11.04DD990 pKa = 3.44KK991 pKa = 9.97TVQVIPEE998 pKa = 4.19DD999 pKa = 3.68QEE1001 pKa = 4.0EE1002 pKa = 4.18LSIEE1006 pKa = 4.1LKK1008 pKa = 9.97IDD1010 pKa = 3.29QVNVIDD1016 pKa = 3.72QDD1018 pKa = 4.27TIISYY1023 pKa = 10.5DD1024 pKa = 3.7DD1025 pKa = 3.83KK1026 pKa = 11.36SAHH1029 pKa = 7.2DD1030 pKa = 4.3EE1031 pKa = 4.55NIHH1034 pKa = 6.64HH1035 pKa = 7.66ADD1037 pKa = 3.26TGIATLWDD1045 pKa = 3.87WIKK1048 pKa = 11.44APFNWVASFFGSFFDD1063 pKa = 3.68LVRR1066 pKa = 11.84VILVILAVIVGLYY1079 pKa = 9.16ILGYY1083 pKa = 9.82IYY1085 pKa = 10.83NLSFSYY1091 pKa = 10.86YY1092 pKa = 9.06KK1093 pKa = 10.32EE1094 pKa = 3.55KK1095 pKa = 10.64RR1096 pKa = 11.84KK1097 pKa = 10.45RR1098 pKa = 11.84KK1099 pKa = 8.85MEE1101 pKa = 4.21FDD1103 pKa = 3.53LEE1105 pKa = 4.14EE1106 pKa = 5.89AEE1108 pKa = 4.13EE1109 pKa = 4.06SLIIKK1114 pKa = 8.8SNKK1117 pKa = 8.26YY1118 pKa = 9.23GEE1120 pKa = 4.27PRR1122 pKa = 11.84RR1123 pKa = 11.84RR1124 pKa = 11.84TPPKK1128 pKa = 9.63TFQFPLDD1135 pKa = 3.67LL1136 pKa = 5.62
MM1 pKa = 7.27NLHH4 pKa = 6.51YY5 pKa = 10.95LLLYY9 pKa = 10.52CFGVYY14 pKa = 10.61LMFTITDD21 pKa = 3.57VLLMNNAEE29 pKa = 4.02DD30 pKa = 4.13HH31 pKa = 6.86EE32 pKa = 4.85DD33 pKa = 2.9IKK35 pKa = 11.1MEE37 pKa = 4.6LKK39 pKa = 10.43RR40 pKa = 11.84LKK42 pKa = 10.42DD43 pKa = 3.27RR44 pKa = 11.84YY45 pKa = 10.87KK46 pKa = 11.03NDD48 pKa = 3.56DD49 pKa = 3.93PEE51 pKa = 5.33DD52 pKa = 4.49LIEE55 pKa = 4.92NKK57 pKa = 10.16VIDD60 pKa = 4.4PGYY63 pKa = 10.13SFPSLDD69 pKa = 3.3PKK71 pKa = 10.53IPEE74 pKa = 4.67RR75 pKa = 11.84IPRR78 pKa = 11.84EE79 pKa = 3.71ATTNVPSTTNVPSTTDD95 pKa = 3.21RR96 pKa = 11.84QVVEE100 pKa = 4.6TLSCDD105 pKa = 3.9DD106 pKa = 4.15FDD108 pKa = 5.4KK109 pKa = 11.17SKK111 pKa = 11.07CIIKK115 pKa = 8.88GTSDD119 pKa = 3.71FNAHH123 pKa = 5.15YY124 pKa = 9.49QVKK127 pKa = 10.5GSDD130 pKa = 3.54SVIACITDD138 pKa = 3.33SEE140 pKa = 4.98KK141 pKa = 10.82IFDD144 pKa = 3.96ACLFDD149 pKa = 5.57SDD151 pKa = 3.42IKK153 pKa = 10.95RR154 pKa = 11.84RR155 pKa = 11.84TFLKK159 pKa = 10.43VPVVPVLKK167 pKa = 10.8LEE169 pKa = 4.2NKK171 pKa = 9.7RR172 pKa = 11.84VLEE175 pKa = 4.18VGSKK179 pKa = 10.11FFFVDD184 pKa = 3.69SSNDD188 pKa = 4.01PILSDD193 pKa = 4.32PKK195 pKa = 10.92SGMANPTVSRR205 pKa = 11.84LSVRR209 pKa = 11.84LSGDD213 pKa = 3.57CIINQVSMSSPYY225 pKa = 9.23MIKK228 pKa = 10.05IKK230 pKa = 9.66STEE233 pKa = 4.17TIGFAVKK240 pKa = 10.41NIKK243 pKa = 10.03NPQAINFQTASGDD256 pKa = 3.08KK257 pKa = 10.33SVYY260 pKa = 10.38FKK262 pKa = 10.52IDD264 pKa = 3.52EE265 pKa = 4.43LDD267 pKa = 3.69GNHH270 pKa = 6.31FFLCGDD276 pKa = 3.18KK277 pKa = 11.08SSFIKK282 pKa = 10.69KK283 pKa = 9.47VDD285 pKa = 3.46VPVRR289 pKa = 11.84NCVSKK294 pKa = 11.27YY295 pKa = 10.19ADD297 pKa = 3.58EE298 pKa = 4.34PKK300 pKa = 10.64KK301 pKa = 10.8IFFCTNFSYY310 pKa = 10.04FKK312 pKa = 10.28WFFTFLVITFPITWILWKK330 pKa = 10.46AKK332 pKa = 9.43NAFSIWYY339 pKa = 8.8DD340 pKa = 2.83IMGILTYY347 pKa = 10.28PILISINYY355 pKa = 8.04LWLYY359 pKa = 10.63FPFKK363 pKa = 10.63CKK365 pKa = 10.49SCGNLSFLSHH375 pKa = 6.22EE376 pKa = 4.38CSRR379 pKa = 11.84LCVCNQSKK387 pKa = 10.24PSRR390 pKa = 11.84EE391 pKa = 3.97HH392 pKa = 7.2SKK394 pKa = 10.57EE395 pKa = 3.73CYY397 pKa = 9.77LFHH400 pKa = 7.89KK401 pKa = 8.97DD402 pKa = 3.14TKK404 pKa = 9.93KK405 pKa = 9.73WRR407 pKa = 11.84SLSLIDD413 pKa = 3.64HH414 pKa = 6.51FQFIVNTKK422 pKa = 9.2ISSEE426 pKa = 3.9FLVFLTKK433 pKa = 9.98MLIAGVLMSCIPSSLALSSEE453 pKa = 4.12RR454 pKa = 11.84NVCVEE459 pKa = 3.55KK460 pKa = 10.91CFYY463 pKa = 11.22SKK465 pKa = 10.9DD466 pKa = 3.79LEE468 pKa = 4.38KK469 pKa = 10.66LTTDD473 pKa = 3.27KK474 pKa = 11.33DD475 pKa = 3.85GSIKK479 pKa = 10.72DD480 pKa = 3.99SIDD483 pKa = 3.2TCEE486 pKa = 4.11CSIGEE491 pKa = 4.81LITEE495 pKa = 4.4TVYY498 pKa = 10.87RR499 pKa = 11.84GGAPISRR506 pKa = 11.84AAIKK510 pKa = 10.14NDD512 pKa = 3.37CVSLSSKK519 pKa = 10.4CLNSSNQAEE528 pKa = 4.21NLFACRR534 pKa = 11.84HH535 pKa = 5.08GCNALSTLNKK545 pKa = 9.16IPKK548 pKa = 9.04VMYY551 pKa = 9.64DD552 pKa = 2.87RR553 pKa = 11.84GYY555 pKa = 11.02KK556 pKa = 10.05GVEE559 pKa = 3.7FSGNLTVLKK568 pKa = 10.16IANRR572 pKa = 11.84LRR574 pKa = 11.84KK575 pKa = 10.13GFVDD579 pKa = 3.45NQSEE583 pKa = 4.35VRR585 pKa = 11.84NLEE588 pKa = 4.19SKK590 pKa = 10.24VSKK593 pKa = 9.78EE594 pKa = 3.46LSYY597 pKa = 11.0FKK599 pKa = 10.56SLKK602 pKa = 10.27VDD604 pKa = 3.77DD605 pKa = 4.73VPPEE609 pKa = 3.92NLMPRR614 pKa = 11.84QSLVFSTEE622 pKa = 3.11VDD624 pKa = 3.05GKK626 pKa = 8.53YY627 pKa = 10.23RR628 pKa = 11.84YY629 pKa = 9.01MIEE632 pKa = 3.79MDD634 pKa = 3.16IKK636 pKa = 10.92KK637 pKa = 8.53EE638 pKa = 4.09TGSVFLLNDD647 pKa = 4.14DD648 pKa = 4.19SSHH651 pKa = 7.66IPMEE655 pKa = 3.59FMVYY659 pKa = 8.85VKK661 pKa = 10.53SVGVEE666 pKa = 3.2YY667 pKa = 10.4DD668 pKa = 2.72IRR670 pKa = 11.84YY671 pKa = 9.5KK672 pKa = 11.09YY673 pKa = 9.68STAKK677 pKa = 10.37VDD679 pKa = 3.74TTVTDD684 pKa = 3.76YY685 pKa = 11.68LSTCTGDD692 pKa = 4.76CKK694 pKa = 10.83DD695 pKa = 3.8CRR697 pKa = 11.84KK698 pKa = 10.47SKK700 pKa = 9.71TPSGHH705 pKa = 6.71NDD707 pKa = 2.85FCIQPTSWWGCEE719 pKa = 3.82EE720 pKa = 4.53LGCLAINEE728 pKa = 4.33GAICGHH734 pKa = 5.88CTNVFDD740 pKa = 6.71LSTLVNVYY748 pKa = 9.59QVVEE752 pKa = 3.77SHH754 pKa = 7.14IIAEE758 pKa = 4.15ICIKK762 pKa = 10.55SLNGYY767 pKa = 8.27DD768 pKa = 3.85CRR770 pKa = 11.84KK771 pKa = 10.01HH772 pKa = 6.79SDD774 pKa = 3.3RR775 pKa = 11.84APIQTDD781 pKa = 4.26YY782 pKa = 10.88YY783 pKa = 9.83QLDD786 pKa = 3.82MTVDD790 pKa = 3.42LHH792 pKa = 7.59NDD794 pKa = 3.21FMSTDD799 pKa = 3.18KK800 pKa = 11.32LFAVTKK806 pKa = 8.66NQKK809 pKa = 9.5ILTGSISDD817 pKa = 4.19LSDD820 pKa = 3.93FSSGAFGHH828 pKa = 5.83PQITVDD834 pKa = 3.79GTPLAVKK841 pKa = 9.2ATISRR846 pKa = 11.84DD847 pKa = 3.22QFTWSCSAVGSKK859 pKa = 10.0SVNIKK864 pKa = 9.92QCGLYY869 pKa = 9.56SYY871 pKa = 11.67NMIYY875 pKa = 10.99ALTQSKK881 pKa = 10.43DD882 pKa = 3.32FSVIDD887 pKa = 3.61EE888 pKa = 4.73EE889 pKa = 4.48NNKK892 pKa = 10.63LYY894 pKa = 9.93MIKK897 pKa = 10.51DD898 pKa = 3.6SLVGKK903 pKa = 9.61LKK905 pKa = 10.92VVIDD909 pKa = 3.75MPKK912 pKa = 10.71EE913 pKa = 4.01MFKK916 pKa = 9.53KK917 pKa = 9.2TPVKK921 pKa = 10.31PILSEE926 pKa = 4.02TKK928 pKa = 9.89IICSGCTKK936 pKa = 10.28CAMGLEE942 pKa = 4.51CVMEE946 pKa = 4.2YY947 pKa = 10.46TSDD950 pKa = 3.36TTFSSRR956 pKa = 11.84LKK958 pKa = 10.11MDD960 pKa = 3.74GCSFKK965 pKa = 10.77SDD967 pKa = 3.6QIGSYY972 pKa = 9.9IGPNKK977 pKa = 10.2KK978 pKa = 10.07LIKK981 pKa = 9.74AYY983 pKa = 10.36CSEE986 pKa = 4.21SVKK989 pKa = 11.04DD990 pKa = 3.44KK991 pKa = 9.97TVQVIPEE998 pKa = 4.19DD999 pKa = 3.68QEE1001 pKa = 4.0EE1002 pKa = 4.18LSIEE1006 pKa = 4.1LKK1008 pKa = 9.97IDD1010 pKa = 3.29QVNVIDD1016 pKa = 3.72QDD1018 pKa = 4.27TIISYY1023 pKa = 10.5DD1024 pKa = 3.7DD1025 pKa = 3.83KK1026 pKa = 11.36SAHH1029 pKa = 7.2DD1030 pKa = 4.3EE1031 pKa = 4.55NIHH1034 pKa = 6.64HH1035 pKa = 7.66ADD1037 pKa = 3.26TGIATLWDD1045 pKa = 3.87WIKK1048 pKa = 11.44APFNWVASFFGSFFDD1063 pKa = 3.68LVRR1066 pKa = 11.84VILVILAVIVGLYY1079 pKa = 9.16ILGYY1083 pKa = 9.82IYY1085 pKa = 10.83NLSFSYY1091 pKa = 10.86YY1092 pKa = 9.06KK1093 pKa = 10.32EE1094 pKa = 3.55KK1095 pKa = 10.64RR1096 pKa = 11.84KK1097 pKa = 10.45RR1098 pKa = 11.84KK1099 pKa = 8.85MEE1101 pKa = 4.21FDD1103 pKa = 3.53LEE1105 pKa = 4.14EE1106 pKa = 5.89AEE1108 pKa = 4.13EE1109 pKa = 4.06SLIIKK1114 pKa = 8.8SNKK1117 pKa = 8.26YY1118 pKa = 9.23GEE1120 pKa = 4.27PRR1122 pKa = 11.84RR1123 pKa = 11.84RR1124 pKa = 11.84TPPKK1128 pKa = 9.63TFQFPLDD1135 pKa = 3.67LL1136 pKa = 5.62
Molecular weight: 128.86 kDa
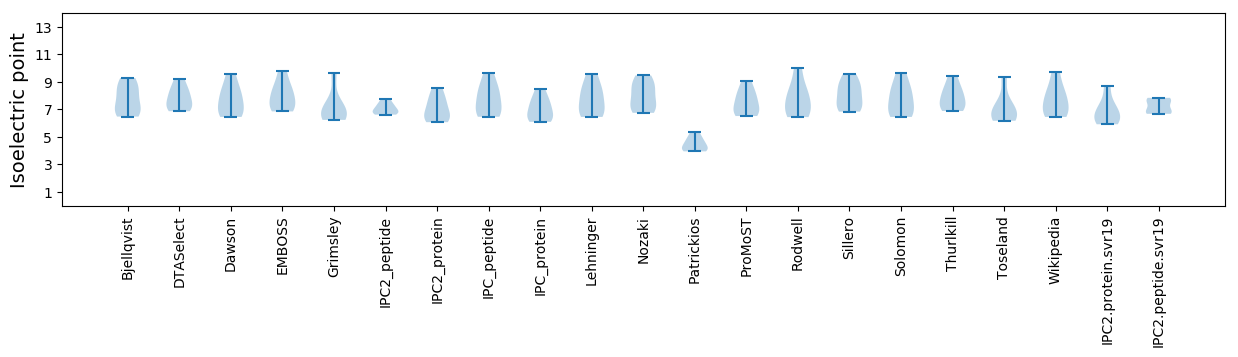
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8V6X1|Q8V6X1_9VIRU Envelopment polyprotein OS=Iris yellow spot virus OX=60456 GN=GP PE=3 SV=1
MM1 pKa = 6.61STVRR5 pKa = 11.84VKK7 pKa = 10.67PSEE10 pKa = 3.94IEE12 pKa = 3.97KK13 pKa = 10.48LLSGGDD19 pKa = 3.19VDD21 pKa = 5.66VVIEE25 pKa = 4.06SDD27 pKa = 3.26EE28 pKa = 4.39TEE30 pKa = 4.08GFNFKK35 pKa = 10.89NFVLANEE42 pKa = 4.8GVQMTFNNGYY52 pKa = 8.29TILRR56 pKa = 11.84NRR58 pKa = 11.84AGIYY62 pKa = 8.65KK63 pKa = 9.27TIKK66 pKa = 8.24TGKK69 pKa = 8.6FTFQGKK75 pKa = 8.15TIVIPSANVSPNQDD89 pKa = 2.3DD90 pKa = 3.58WTFRR94 pKa = 11.84RR95 pKa = 11.84LEE97 pKa = 3.93GFIRR101 pKa = 11.84ARR103 pKa = 11.84MLVEE107 pKa = 4.84LIEE110 pKa = 4.45TKK112 pKa = 10.46DD113 pKa = 3.46EE114 pKa = 4.15KK115 pKa = 10.84EE116 pKa = 3.9KK117 pKa = 11.02QKK119 pKa = 10.27MYY121 pKa = 11.02EE122 pKa = 4.31KK123 pKa = 10.3ICGLPLVSAYY133 pKa = 10.15GLKK136 pKa = 10.34PSSKK140 pKa = 9.73FHH142 pKa = 6.3ATTARR147 pKa = 11.84IMLTLGGPLTLMASLDD163 pKa = 3.54IFAAAALPLAYY174 pKa = 9.83FQNVKK179 pKa = 10.5KK180 pKa = 9.87EE181 pKa = 3.81ALGISRR187 pKa = 11.84FSTYY191 pKa = 9.83EE192 pKa = 3.58QLCKK196 pKa = 10.03VARR199 pKa = 11.84VMAAKK204 pKa = 9.95EE205 pKa = 4.04FKK207 pKa = 9.89FTEE210 pKa = 4.36KK211 pKa = 10.36YY212 pKa = 10.58KK213 pKa = 10.91KK214 pKa = 10.44IFDD217 pKa = 3.76EE218 pKa = 4.78TIKK221 pKa = 10.65ILTDD225 pKa = 3.56CTPGTSGAASLIKK238 pKa = 10.52FNEE241 pKa = 3.98QIKK244 pKa = 10.11ILEE247 pKa = 4.29GAFGKK252 pKa = 10.12IVEE255 pKa = 5.34DD256 pKa = 3.22IGEE259 pKa = 4.16SSKK262 pKa = 11.19PKK264 pKa = 9.98TPSKK268 pKa = 10.28KK269 pKa = 10.0DD270 pKa = 3.08RR271 pKa = 11.84YY272 pKa = 9.62NN273 pKa = 3.03
MM1 pKa = 6.61STVRR5 pKa = 11.84VKK7 pKa = 10.67PSEE10 pKa = 3.94IEE12 pKa = 3.97KK13 pKa = 10.48LLSGGDD19 pKa = 3.19VDD21 pKa = 5.66VVIEE25 pKa = 4.06SDD27 pKa = 3.26EE28 pKa = 4.39TEE30 pKa = 4.08GFNFKK35 pKa = 10.89NFVLANEE42 pKa = 4.8GVQMTFNNGYY52 pKa = 8.29TILRR56 pKa = 11.84NRR58 pKa = 11.84AGIYY62 pKa = 8.65KK63 pKa = 9.27TIKK66 pKa = 8.24TGKK69 pKa = 8.6FTFQGKK75 pKa = 8.15TIVIPSANVSPNQDD89 pKa = 2.3DD90 pKa = 3.58WTFRR94 pKa = 11.84RR95 pKa = 11.84LEE97 pKa = 3.93GFIRR101 pKa = 11.84ARR103 pKa = 11.84MLVEE107 pKa = 4.84LIEE110 pKa = 4.45TKK112 pKa = 10.46DD113 pKa = 3.46EE114 pKa = 4.15KK115 pKa = 10.84EE116 pKa = 3.9KK117 pKa = 11.02QKK119 pKa = 10.27MYY121 pKa = 11.02EE122 pKa = 4.31KK123 pKa = 10.3ICGLPLVSAYY133 pKa = 10.15GLKK136 pKa = 10.34PSSKK140 pKa = 9.73FHH142 pKa = 6.3ATTARR147 pKa = 11.84IMLTLGGPLTLMASLDD163 pKa = 3.54IFAAAALPLAYY174 pKa = 9.83FQNVKK179 pKa = 10.5KK180 pKa = 9.87EE181 pKa = 3.81ALGISRR187 pKa = 11.84FSTYY191 pKa = 9.83EE192 pKa = 3.58QLCKK196 pKa = 10.03VARR199 pKa = 11.84VMAAKK204 pKa = 9.95EE205 pKa = 4.04FKK207 pKa = 9.89FTEE210 pKa = 4.36KK211 pKa = 10.36YY212 pKa = 10.58KK213 pKa = 10.91KK214 pKa = 10.44IFDD217 pKa = 3.76EE218 pKa = 4.78TIKK221 pKa = 10.65ILTDD225 pKa = 3.56CTPGTSGAASLIKK238 pKa = 10.52FNEE241 pKa = 3.98QIKK244 pKa = 10.11ILEE247 pKa = 4.29GAFGKK252 pKa = 10.12IVEE255 pKa = 5.34DD256 pKa = 3.22IGEE259 pKa = 4.16SSKK262 pKa = 11.19PKK264 pKa = 9.98TPSKK268 pKa = 10.28KK269 pKa = 10.0DD270 pKa = 3.08RR271 pKa = 11.84YY272 pKa = 9.62NN273 pKa = 3.03
Molecular weight: 30.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6483 |
273 |
2873 |
926.1 |
105.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.795 ± 0.529 | 2.453 ± 0.373 |
6.355 ± 0.393 | 6.185 ± 0.383 |
4.936 ± 0.24 | 4.257 ± 0.356 |
1.512 ± 0.146 | 7.605 ± 0.16 |
9.27 ± 0.206 | 8.808 ± 0.612 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.178 ± 0.538 | 6.047 ± 0.6 |
3.039 ± 0.384 | 2.391 ± 0.09 |
3.378 ± 0.095 | 9.918 ± 0.281 |
5.954 ± 0.205 | 6.016 ± 0.393 |
0.848 ± 0.126 | 4.057 ± 0.34 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
