
Mangrovibacterium diazotrophicum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Prolixibacteraceae; Mangrovibacterium
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
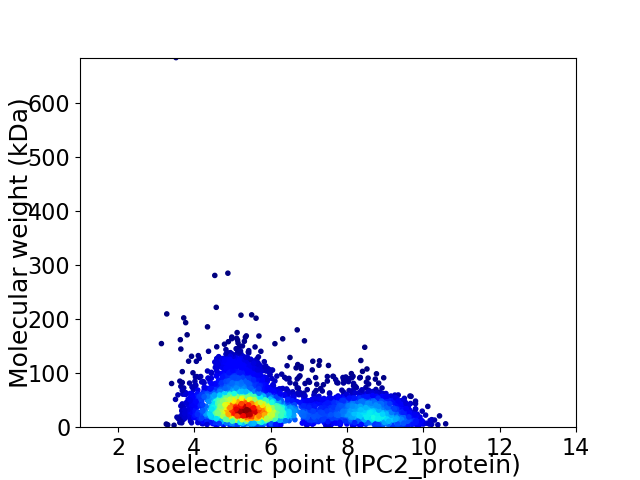
Virtual 2D-PAGE plot for 4641 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
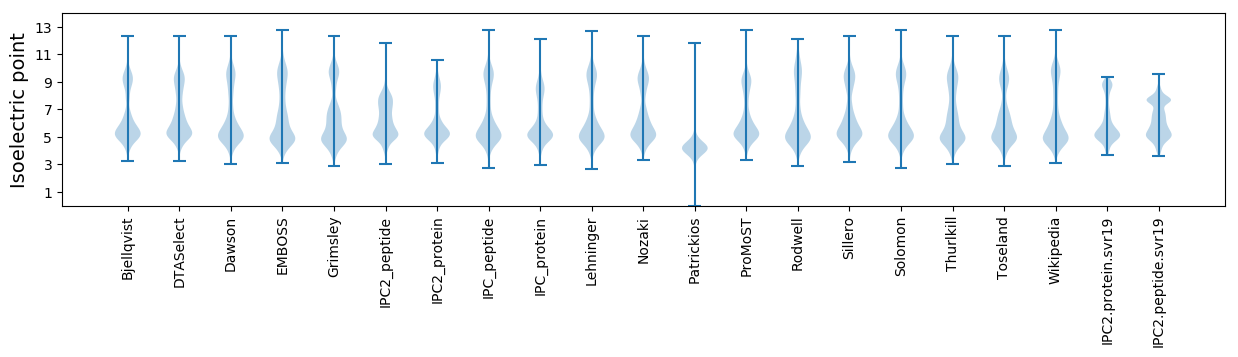
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A419VY99|A0A419VY99_9BACT Signal recognition particle protein OS=Mangrovibacterium diazotrophicum OX=1261403 GN=ffh PE=3 SV=1
MM1 pKa = 7.48NNPTTHH7 pKa = 5.78FMKK10 pKa = 10.59VNHH13 pKa = 6.81KK14 pKa = 10.53LLILSLSIGLMTALSCKK31 pKa = 10.33KK32 pKa = 10.43YY33 pKa = 10.95DD34 pKa = 4.88GDD36 pKa = 6.01DD37 pKa = 3.73SDD39 pKa = 6.43DD40 pKa = 5.38DD41 pKa = 5.22DD42 pKa = 7.11DD43 pKa = 3.87EE44 pKa = 4.58TTIVDD49 pKa = 3.93AEE51 pKa = 4.25TGNAEE56 pKa = 4.13DD57 pKa = 4.46HH58 pKa = 6.82EE59 pKa = 4.53EE60 pKa = 4.0ASDD63 pKa = 4.05YY64 pKa = 10.91VWEE67 pKa = 5.14GDD69 pKa = 3.63TVTAIILNEE78 pKa = 3.84TSITVSGDD86 pKa = 2.94GASASGTTATVTAAGTYY103 pKa = 9.5LVSGTLDD110 pKa = 3.12NGKK113 pKa = 10.08IIVDD117 pKa = 4.06TEE119 pKa = 4.24DD120 pKa = 3.23EE121 pKa = 4.39DD122 pKa = 4.66AVKK125 pKa = 10.64LVLNAVNITCSNSAPIFVKK144 pKa = 10.46SAEE147 pKa = 4.07KK148 pKa = 10.49AIVILPSGTTNYY160 pKa = 8.93LTDD163 pKa = 4.11GSSYY167 pKa = 10.94SYY169 pKa = 11.05SSSDD173 pKa = 3.14EE174 pKa = 4.4DD175 pKa = 3.72EE176 pKa = 4.79PNAALYY182 pKa = 10.6SKK184 pKa = 10.89SDD186 pKa = 3.53LSISGEE192 pKa = 3.99GALVVDD198 pKa = 4.85ANYY201 pKa = 11.03NDD203 pKa = 5.37GITSKK208 pKa = 10.79DD209 pKa = 3.32GLIIASGNISVNSVDD224 pKa = 4.3DD225 pKa = 4.77GIRR228 pKa = 11.84GKK230 pKa = 10.4DD231 pKa = 3.41YY232 pKa = 11.33LIIEE236 pKa = 4.68SGTINVTSGGDD247 pKa = 3.56GFKK250 pKa = 10.7SDD252 pKa = 4.99NDD254 pKa = 3.25EE255 pKa = 4.91DD256 pKa = 4.13DD257 pKa = 3.73AKK259 pKa = 11.19GYY261 pKa = 10.46ISIEE265 pKa = 4.16SGTITITAANDD276 pKa = 3.21GMQATTDD283 pKa = 3.91LLIADD288 pKa = 4.05GTFNLTTGGGSSKK301 pKa = 9.69TISSSVSAKK310 pKa = 10.23GLKK313 pKa = 9.74SDD315 pKa = 4.03LNIIIEE321 pKa = 4.52SGDD324 pKa = 3.45FTINSADD331 pKa = 3.9DD332 pKa = 5.08AIHH335 pKa = 5.93SASLITINDD344 pKa = 3.41GTIAISSGDD353 pKa = 3.92DD354 pKa = 4.37GIHH357 pKa = 6.6TDD359 pKa = 2.93NTVNVNGGEE368 pKa = 3.92ITITKK373 pKa = 9.62SYY375 pKa = 10.67EE376 pKa = 4.45GIEE379 pKa = 4.17GPYY382 pKa = 10.35INMNGGTTYY391 pKa = 9.28VTSSDD396 pKa = 3.87DD397 pKa = 3.46GLNASKK403 pKa = 11.33GNGGEE408 pKa = 4.31SNDD411 pKa = 3.6GSLLSITGGYY421 pKa = 10.23LYY423 pKa = 10.21ISSTGDD429 pKa = 3.34GLDD432 pKa = 3.57SNGSITSTGGTVIVNGPSSSVEE454 pKa = 3.63VGLDD458 pKa = 3.46YY459 pKa = 11.63NGTFSLNGGLILIAGPNSSMTQGASSSSTQNSVLVRR495 pKa = 11.84FSSSKK500 pKa = 10.51SSSTLFNIQDD510 pKa = 3.25ASGNSLVTFQPAKK523 pKa = 8.98TYY525 pKa = 11.16SSVLYY530 pKa = 10.56SSSDD534 pKa = 3.36LTKK537 pKa = 9.08GTTYY541 pKa = 10.69YY542 pKa = 9.81IYY544 pKa = 10.95SGGSYY549 pKa = 10.28SGGSEE554 pKa = 3.83TDD556 pKa = 3.13GLYY559 pKa = 10.32TGGTYY564 pKa = 10.33SGGTQYY570 pKa = 11.67SSFTVSGSVTSVGSSSGSSGGGGGGRR596 pKa = 3.79
MM1 pKa = 7.48NNPTTHH7 pKa = 5.78FMKK10 pKa = 10.59VNHH13 pKa = 6.81KK14 pKa = 10.53LLILSLSIGLMTALSCKK31 pKa = 10.33KK32 pKa = 10.43YY33 pKa = 10.95DD34 pKa = 4.88GDD36 pKa = 6.01DD37 pKa = 3.73SDD39 pKa = 6.43DD40 pKa = 5.38DD41 pKa = 5.22DD42 pKa = 7.11DD43 pKa = 3.87EE44 pKa = 4.58TTIVDD49 pKa = 3.93AEE51 pKa = 4.25TGNAEE56 pKa = 4.13DD57 pKa = 4.46HH58 pKa = 6.82EE59 pKa = 4.53EE60 pKa = 4.0ASDD63 pKa = 4.05YY64 pKa = 10.91VWEE67 pKa = 5.14GDD69 pKa = 3.63TVTAIILNEE78 pKa = 3.84TSITVSGDD86 pKa = 2.94GASASGTTATVTAAGTYY103 pKa = 9.5LVSGTLDD110 pKa = 3.12NGKK113 pKa = 10.08IIVDD117 pKa = 4.06TEE119 pKa = 4.24DD120 pKa = 3.23EE121 pKa = 4.39DD122 pKa = 4.66AVKK125 pKa = 10.64LVLNAVNITCSNSAPIFVKK144 pKa = 10.46SAEE147 pKa = 4.07KK148 pKa = 10.49AIVILPSGTTNYY160 pKa = 8.93LTDD163 pKa = 4.11GSSYY167 pKa = 10.94SYY169 pKa = 11.05SSSDD173 pKa = 3.14EE174 pKa = 4.4DD175 pKa = 3.72EE176 pKa = 4.79PNAALYY182 pKa = 10.6SKK184 pKa = 10.89SDD186 pKa = 3.53LSISGEE192 pKa = 3.99GALVVDD198 pKa = 4.85ANYY201 pKa = 11.03NDD203 pKa = 5.37GITSKK208 pKa = 10.79DD209 pKa = 3.32GLIIASGNISVNSVDD224 pKa = 4.3DD225 pKa = 4.77GIRR228 pKa = 11.84GKK230 pKa = 10.4DD231 pKa = 3.41YY232 pKa = 11.33LIIEE236 pKa = 4.68SGTINVTSGGDD247 pKa = 3.56GFKK250 pKa = 10.7SDD252 pKa = 4.99NDD254 pKa = 3.25EE255 pKa = 4.91DD256 pKa = 4.13DD257 pKa = 3.73AKK259 pKa = 11.19GYY261 pKa = 10.46ISIEE265 pKa = 4.16SGTITITAANDD276 pKa = 3.21GMQATTDD283 pKa = 3.91LLIADD288 pKa = 4.05GTFNLTTGGGSSKK301 pKa = 9.69TISSSVSAKK310 pKa = 10.23GLKK313 pKa = 9.74SDD315 pKa = 4.03LNIIIEE321 pKa = 4.52SGDD324 pKa = 3.45FTINSADD331 pKa = 3.9DD332 pKa = 5.08AIHH335 pKa = 5.93SASLITINDD344 pKa = 3.41GTIAISSGDD353 pKa = 3.92DD354 pKa = 4.37GIHH357 pKa = 6.6TDD359 pKa = 2.93NTVNVNGGEE368 pKa = 3.92ITITKK373 pKa = 9.62SYY375 pKa = 10.67EE376 pKa = 4.45GIEE379 pKa = 4.17GPYY382 pKa = 10.35INMNGGTTYY391 pKa = 9.28VTSSDD396 pKa = 3.87DD397 pKa = 3.46GLNASKK403 pKa = 11.33GNGGEE408 pKa = 4.31SNDD411 pKa = 3.6GSLLSITGGYY421 pKa = 10.23LYY423 pKa = 10.21ISSTGDD429 pKa = 3.34GLDD432 pKa = 3.57SNGSITSTGGTVIVNGPSSSVEE454 pKa = 3.63VGLDD458 pKa = 3.46YY459 pKa = 11.63NGTFSLNGGLILIAGPNSSMTQGASSSSTQNSVLVRR495 pKa = 11.84FSSSKK500 pKa = 10.51SSSTLFNIQDD510 pKa = 3.25ASGNSLVTFQPAKK523 pKa = 8.98TYY525 pKa = 11.16SSVLYY530 pKa = 10.56SSSDD534 pKa = 3.36LTKK537 pKa = 9.08GTTYY541 pKa = 10.69YY542 pKa = 9.81IYY544 pKa = 10.95SGGSYY549 pKa = 10.28SGGSEE554 pKa = 3.83TDD556 pKa = 3.13GLYY559 pKa = 10.32TGGTYY564 pKa = 10.33SGGTQYY570 pKa = 11.67SSFTVSGSVTSVGSSSGSSGGGGGGRR596 pKa = 3.79
Molecular weight: 60.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A419VVZ8|A0A419VVZ8_9BACT Iron-only hydrogenase maturation protein HydG OS=Mangrovibacterium diazotrophicum OX=1261403 GN=BC643_4014 PE=4 SV=1
MM1 pKa = 7.28AQIKK5 pKa = 9.38QRR7 pKa = 11.84GVIEE11 pKa = 4.33LGKK14 pKa = 10.04SLLMVVLGLALLFIPEE30 pKa = 4.05VTLVSLARR38 pKa = 11.84LVAIFLIVQGIFVALSAFRR57 pKa = 11.84YY58 pKa = 9.35RR59 pKa = 11.84RR60 pKa = 11.84ADD62 pKa = 3.01INKK65 pKa = 8.97YY66 pKa = 10.85AFGLEE71 pKa = 4.12AGVSLIVGFIILYY84 pKa = 10.41NPGTTISFFVVVLAIWAILGGLLMAFSFNQLRR116 pKa = 11.84KK117 pKa = 10.11AGVTSWFLFFNSLVAMTFGLVLIFEE142 pKa = 4.68PLRR145 pKa = 11.84GGFALASIIGAFVLLHH161 pKa = 6.29GLLSLFSAFSRR172 pKa = 3.95
MM1 pKa = 7.28AQIKK5 pKa = 9.38QRR7 pKa = 11.84GVIEE11 pKa = 4.33LGKK14 pKa = 10.04SLLMVVLGLALLFIPEE30 pKa = 4.05VTLVSLARR38 pKa = 11.84LVAIFLIVQGIFVALSAFRR57 pKa = 11.84YY58 pKa = 9.35RR59 pKa = 11.84RR60 pKa = 11.84ADD62 pKa = 3.01INKK65 pKa = 8.97YY66 pKa = 10.85AFGLEE71 pKa = 4.12AGVSLIVGFIILYY84 pKa = 10.41NPGTTISFFVVVLAIWAILGGLLMAFSFNQLRR116 pKa = 11.84KK117 pKa = 10.11AGVTSWFLFFNSLVAMTFGLVLIFEE142 pKa = 4.68PLRR145 pKa = 11.84GGFALASIIGAFVLLHH161 pKa = 6.29GLLSLFSAFSRR172 pKa = 3.95
Molecular weight: 18.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1732579 |
30 |
6730 |
373.3 |
41.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.032 ± 0.036 | 0.883 ± 0.016 |
5.818 ± 0.025 | 6.625 ± 0.032 |
4.991 ± 0.026 | 6.862 ± 0.031 |
1.744 ± 0.019 | 6.843 ± 0.032 |
6.444 ± 0.038 | 9.378 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.412 ± 0.019 | 5.206 ± 0.028 |
3.747 ± 0.021 | 3.699 ± 0.021 |
4.037 ± 0.027 | 6.634 ± 0.04 |
5.518 ± 0.057 | 6.593 ± 0.033 |
1.328 ± 0.015 | 4.204 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
