
Sanxia water strider virus 15
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 9.03
Get precalculated fractions of proteins
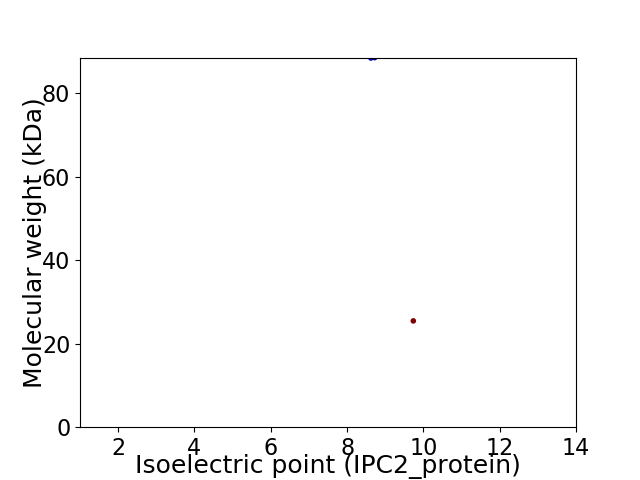
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG98|A0A1L3KG98_9VIRU RNA-directed RNA polymerase OS=Sanxia water strider virus 15 OX=1923399 PE=4 SV=1
MM1 pKa = 7.51SNYY4 pKa = 10.21AINILEE10 pKa = 4.44CNSLPSSPVGHH21 pKa = 6.54MEE23 pKa = 3.56TAKK26 pKa = 10.22WYY28 pKa = 9.92DD29 pKa = 3.56CMRR32 pKa = 11.84MFRR35 pKa = 11.84APRR38 pKa = 11.84RR39 pKa = 11.84WTRR42 pKa = 11.84SLKK45 pKa = 10.85APAAPKK51 pKa = 8.58WVKK54 pKa = 10.34PKK56 pKa = 9.78WSSLAKK62 pKa = 9.33PFKK65 pKa = 10.1RR66 pKa = 11.84VKK68 pKa = 8.91TPKK71 pKa = 9.93RR72 pKa = 11.84EE73 pKa = 3.97PGKK76 pKa = 9.22VTEE79 pKa = 4.27EE80 pKa = 3.99SPYY83 pKa = 10.0PANLSPEE90 pKa = 3.91SRR92 pKa = 11.84TWLTANPTGVTAAIRR107 pKa = 11.84VLNAPSKK114 pKa = 10.19DD115 pKa = 3.59AQPKK119 pKa = 7.61YY120 pKa = 10.61LKK122 pKa = 10.32SALMYY127 pKa = 10.73DD128 pKa = 4.31RR129 pKa = 11.84LCIGHH134 pKa = 7.32DD135 pKa = 3.2RR136 pKa = 11.84LIPNTTEE143 pKa = 3.7FRR145 pKa = 11.84WPNEE149 pKa = 3.57RR150 pKa = 11.84VRR152 pKa = 11.84LCGLIPVHH160 pKa = 7.19RR161 pKa = 11.84IAKK164 pKa = 8.53LHH166 pKa = 4.82NHH168 pKa = 5.05KK169 pKa = 10.48HH170 pKa = 5.5EE171 pKa = 5.14DD172 pKa = 3.14LYY174 pKa = 11.23QYY176 pKa = 11.38LRR178 pKa = 11.84IKK180 pKa = 10.64SMYY183 pKa = 10.8AMRR186 pKa = 11.84SPLLLKK192 pKa = 9.84QLKK195 pKa = 8.97QHH197 pKa = 6.51ALQWLEE203 pKa = 3.94GFDD206 pKa = 3.54TSEE209 pKa = 3.86YY210 pKa = 9.91TPAGLADD217 pKa = 5.43LISRR221 pKa = 11.84TCAQAYY227 pKa = 10.28AGDD230 pKa = 4.17AFDD233 pKa = 4.5ITLEE237 pKa = 4.07TYY239 pKa = 8.51MAEE242 pKa = 4.11RR243 pKa = 11.84QGLHH247 pKa = 6.51PSPSSFHH254 pKa = 6.14HH255 pKa = 6.79RR256 pKa = 11.84PYY258 pKa = 9.99SWRR261 pKa = 11.84RR262 pKa = 11.84VYY264 pKa = 10.96AWLRR268 pKa = 11.84IMKK271 pKa = 10.03LEE273 pKa = 4.19LSSAVFKK280 pKa = 10.67RR281 pKa = 11.84IPPLVCNNRR290 pKa = 11.84TMYY293 pKa = 11.12LDD295 pKa = 3.73LHH297 pKa = 5.99SHH299 pKa = 5.36SFGYY303 pKa = 10.03RR304 pKa = 11.84VGLHH308 pKa = 6.12SSCICNEE315 pKa = 4.03IIAISNRR322 pKa = 11.84HH323 pKa = 6.17CIEE326 pKa = 3.88NPLRR330 pKa = 11.84TSRR333 pKa = 11.84QFDD336 pKa = 3.73VYY338 pKa = 11.19AEE340 pKa = 4.06EE341 pKa = 4.23SVKK344 pKa = 10.5FFNFTIQKK352 pKa = 8.91ISKK355 pKa = 8.65EE356 pKa = 4.23EE357 pKa = 3.94VISRR361 pKa = 11.84YY362 pKa = 8.51TGLKK366 pKa = 7.78KK367 pKa = 10.33SRR369 pKa = 11.84YY370 pKa = 8.54IAACTTLRR378 pKa = 11.84EE379 pKa = 4.27QGWHH383 pKa = 6.53KK384 pKa = 10.74FNTHH388 pKa = 5.38ISSFVKK394 pKa = 10.18VDD396 pKa = 3.13KK397 pKa = 11.06APYY400 pKa = 9.89GKK402 pKa = 10.45LFTKK406 pKa = 10.4APRR409 pKa = 11.84LIQGRR414 pKa = 11.84TPEE417 pKa = 4.07YY418 pKa = 10.03NLCYY422 pKa = 9.62LQYY425 pKa = 9.96VVPMEE430 pKa = 3.9HH431 pKa = 6.12TLYY434 pKa = 11.11EE435 pKa = 4.05NLTYY439 pKa = 10.66GSHH442 pKa = 6.36SNLRR446 pKa = 11.84CVAKK450 pKa = 10.45GLNPQEE456 pKa = 4.1RR457 pKa = 11.84AEE459 pKa = 4.41LYY461 pKa = 9.61LGKK464 pKa = 10.01VASYY468 pKa = 10.06IDD470 pKa = 3.61PVVYY474 pKa = 10.72SGDD477 pKa = 3.43HH478 pKa = 5.16KK479 pKa = 10.03TFDD482 pKa = 3.44AFITVHH488 pKa = 5.96HH489 pKa = 6.9LKK491 pKa = 10.42AVHH494 pKa = 5.8RR495 pKa = 11.84KK496 pKa = 6.62YY497 pKa = 11.04RR498 pKa = 11.84KK499 pKa = 7.64MCGRR503 pKa = 11.84GIKK506 pKa = 9.83RR507 pKa = 11.84YY508 pKa = 9.64CHH510 pKa = 6.14AQLHH514 pKa = 5.76NKK516 pKa = 9.15CKK518 pKa = 8.48TVNGIRR524 pKa = 11.84YY525 pKa = 8.0NIQGTRR531 pKa = 11.84MSGDD535 pKa = 3.49ADD537 pKa = 3.55TGLGNTIISLDD548 pKa = 3.54LAYY551 pKa = 9.37ATLRR555 pKa = 11.84SNNIEE560 pKa = 4.77KK561 pKa = 10.72YY562 pKa = 10.59DD563 pKa = 4.16LLCDD567 pKa = 4.19GDD569 pKa = 4.27DD570 pKa = 3.62FMIIVEE576 pKa = 4.78RR577 pKa = 11.84GTILKK582 pKa = 10.61GDD584 pKa = 3.58MFLPFGFRR592 pKa = 11.84TEE594 pKa = 4.15LTQTEE599 pKa = 3.97VDD601 pKa = 3.3MVEE604 pKa = 4.66FCQCRR609 pKa = 11.84IVDD612 pKa = 3.48TGTRR616 pKa = 11.84QVFVRR621 pKa = 11.84NPEE624 pKa = 3.92RR625 pKa = 11.84MLSNSRR631 pKa = 11.84ICRR634 pKa = 11.84KK635 pKa = 10.09AYY637 pKa = 8.58TVQLYY642 pKa = 10.04KK643 pKa = 10.81EE644 pKa = 4.2WLNSVGLCEE653 pKa = 3.77ATLYY657 pKa = 11.26GDD659 pKa = 3.26IPIYY663 pKa = 9.74STYY666 pKa = 10.4SWSQITDD673 pKa = 3.18RR674 pKa = 11.84TACIKK679 pKa = 11.0DD680 pKa = 3.33PDD682 pKa = 3.68MLRR685 pKa = 11.84RR686 pKa = 11.84MEE688 pKa = 4.43GMPIKK693 pKa = 10.5PIDD696 pKa = 3.72TPVTDD701 pKa = 3.63EE702 pKa = 4.65ARR704 pKa = 11.84VSFYY708 pKa = 10.84KK709 pKa = 10.6VWGVPPEE716 pKa = 3.89IQIEE720 pKa = 4.36LEE722 pKa = 4.18SQLTCTNIFSKK733 pKa = 10.05RR734 pKa = 11.84TIKK737 pKa = 10.4SRR739 pKa = 11.84YY740 pKa = 8.32GPCSGQKK747 pKa = 8.54EE748 pKa = 4.14QPLAEE753 pKa = 4.73LSCSSWWSRR762 pKa = 11.84CEE764 pKa = 3.97CGHH767 pKa = 6.75
MM1 pKa = 7.51SNYY4 pKa = 10.21AINILEE10 pKa = 4.44CNSLPSSPVGHH21 pKa = 6.54MEE23 pKa = 3.56TAKK26 pKa = 10.22WYY28 pKa = 9.92DD29 pKa = 3.56CMRR32 pKa = 11.84MFRR35 pKa = 11.84APRR38 pKa = 11.84RR39 pKa = 11.84WTRR42 pKa = 11.84SLKK45 pKa = 10.85APAAPKK51 pKa = 8.58WVKK54 pKa = 10.34PKK56 pKa = 9.78WSSLAKK62 pKa = 9.33PFKK65 pKa = 10.1RR66 pKa = 11.84VKK68 pKa = 8.91TPKK71 pKa = 9.93RR72 pKa = 11.84EE73 pKa = 3.97PGKK76 pKa = 9.22VTEE79 pKa = 4.27EE80 pKa = 3.99SPYY83 pKa = 10.0PANLSPEE90 pKa = 3.91SRR92 pKa = 11.84TWLTANPTGVTAAIRR107 pKa = 11.84VLNAPSKK114 pKa = 10.19DD115 pKa = 3.59AQPKK119 pKa = 7.61YY120 pKa = 10.61LKK122 pKa = 10.32SALMYY127 pKa = 10.73DD128 pKa = 4.31RR129 pKa = 11.84LCIGHH134 pKa = 7.32DD135 pKa = 3.2RR136 pKa = 11.84LIPNTTEE143 pKa = 3.7FRR145 pKa = 11.84WPNEE149 pKa = 3.57RR150 pKa = 11.84VRR152 pKa = 11.84LCGLIPVHH160 pKa = 7.19RR161 pKa = 11.84IAKK164 pKa = 8.53LHH166 pKa = 4.82NHH168 pKa = 5.05KK169 pKa = 10.48HH170 pKa = 5.5EE171 pKa = 5.14DD172 pKa = 3.14LYY174 pKa = 11.23QYY176 pKa = 11.38LRR178 pKa = 11.84IKK180 pKa = 10.64SMYY183 pKa = 10.8AMRR186 pKa = 11.84SPLLLKK192 pKa = 9.84QLKK195 pKa = 8.97QHH197 pKa = 6.51ALQWLEE203 pKa = 3.94GFDD206 pKa = 3.54TSEE209 pKa = 3.86YY210 pKa = 9.91TPAGLADD217 pKa = 5.43LISRR221 pKa = 11.84TCAQAYY227 pKa = 10.28AGDD230 pKa = 4.17AFDD233 pKa = 4.5ITLEE237 pKa = 4.07TYY239 pKa = 8.51MAEE242 pKa = 4.11RR243 pKa = 11.84QGLHH247 pKa = 6.51PSPSSFHH254 pKa = 6.14HH255 pKa = 6.79RR256 pKa = 11.84PYY258 pKa = 9.99SWRR261 pKa = 11.84RR262 pKa = 11.84VYY264 pKa = 10.96AWLRR268 pKa = 11.84IMKK271 pKa = 10.03LEE273 pKa = 4.19LSSAVFKK280 pKa = 10.67RR281 pKa = 11.84IPPLVCNNRR290 pKa = 11.84TMYY293 pKa = 11.12LDD295 pKa = 3.73LHH297 pKa = 5.99SHH299 pKa = 5.36SFGYY303 pKa = 10.03RR304 pKa = 11.84VGLHH308 pKa = 6.12SSCICNEE315 pKa = 4.03IIAISNRR322 pKa = 11.84HH323 pKa = 6.17CIEE326 pKa = 3.88NPLRR330 pKa = 11.84TSRR333 pKa = 11.84QFDD336 pKa = 3.73VYY338 pKa = 11.19AEE340 pKa = 4.06EE341 pKa = 4.23SVKK344 pKa = 10.5FFNFTIQKK352 pKa = 8.91ISKK355 pKa = 8.65EE356 pKa = 4.23EE357 pKa = 3.94VISRR361 pKa = 11.84YY362 pKa = 8.51TGLKK366 pKa = 7.78KK367 pKa = 10.33SRR369 pKa = 11.84YY370 pKa = 8.54IAACTTLRR378 pKa = 11.84EE379 pKa = 4.27QGWHH383 pKa = 6.53KK384 pKa = 10.74FNTHH388 pKa = 5.38ISSFVKK394 pKa = 10.18VDD396 pKa = 3.13KK397 pKa = 11.06APYY400 pKa = 9.89GKK402 pKa = 10.45LFTKK406 pKa = 10.4APRR409 pKa = 11.84LIQGRR414 pKa = 11.84TPEE417 pKa = 4.07YY418 pKa = 10.03NLCYY422 pKa = 9.62LQYY425 pKa = 9.96VVPMEE430 pKa = 3.9HH431 pKa = 6.12TLYY434 pKa = 11.11EE435 pKa = 4.05NLTYY439 pKa = 10.66GSHH442 pKa = 6.36SNLRR446 pKa = 11.84CVAKK450 pKa = 10.45GLNPQEE456 pKa = 4.1RR457 pKa = 11.84AEE459 pKa = 4.41LYY461 pKa = 9.61LGKK464 pKa = 10.01VASYY468 pKa = 10.06IDD470 pKa = 3.61PVVYY474 pKa = 10.72SGDD477 pKa = 3.43HH478 pKa = 5.16KK479 pKa = 10.03TFDD482 pKa = 3.44AFITVHH488 pKa = 5.96HH489 pKa = 6.9LKK491 pKa = 10.42AVHH494 pKa = 5.8RR495 pKa = 11.84KK496 pKa = 6.62YY497 pKa = 11.04RR498 pKa = 11.84KK499 pKa = 7.64MCGRR503 pKa = 11.84GIKK506 pKa = 9.83RR507 pKa = 11.84YY508 pKa = 9.64CHH510 pKa = 6.14AQLHH514 pKa = 5.76NKK516 pKa = 9.15CKK518 pKa = 8.48TVNGIRR524 pKa = 11.84YY525 pKa = 8.0NIQGTRR531 pKa = 11.84MSGDD535 pKa = 3.49ADD537 pKa = 3.55TGLGNTIISLDD548 pKa = 3.54LAYY551 pKa = 9.37ATLRR555 pKa = 11.84SNNIEE560 pKa = 4.77KK561 pKa = 10.72YY562 pKa = 10.59DD563 pKa = 4.16LLCDD567 pKa = 4.19GDD569 pKa = 4.27DD570 pKa = 3.62FMIIVEE576 pKa = 4.78RR577 pKa = 11.84GTILKK582 pKa = 10.61GDD584 pKa = 3.58MFLPFGFRR592 pKa = 11.84TEE594 pKa = 4.15LTQTEE599 pKa = 3.97VDD601 pKa = 3.3MVEE604 pKa = 4.66FCQCRR609 pKa = 11.84IVDD612 pKa = 3.48TGTRR616 pKa = 11.84QVFVRR621 pKa = 11.84NPEE624 pKa = 3.92RR625 pKa = 11.84MLSNSRR631 pKa = 11.84ICRR634 pKa = 11.84KK635 pKa = 10.09AYY637 pKa = 8.58TVQLYY642 pKa = 10.04KK643 pKa = 10.81EE644 pKa = 4.2WLNSVGLCEE653 pKa = 3.77ATLYY657 pKa = 11.26GDD659 pKa = 3.26IPIYY663 pKa = 9.74STYY666 pKa = 10.4SWSQITDD673 pKa = 3.18RR674 pKa = 11.84TACIKK679 pKa = 11.0DD680 pKa = 3.33PDD682 pKa = 3.68MLRR685 pKa = 11.84RR686 pKa = 11.84MEE688 pKa = 4.43GMPIKK693 pKa = 10.5PIDD696 pKa = 3.72TPVTDD701 pKa = 3.63EE702 pKa = 4.65ARR704 pKa = 11.84VSFYY708 pKa = 10.84KK709 pKa = 10.6VWGVPPEE716 pKa = 3.89IQIEE720 pKa = 4.36LEE722 pKa = 4.18SQLTCTNIFSKK733 pKa = 10.05RR734 pKa = 11.84TIKK737 pKa = 10.4SRR739 pKa = 11.84YY740 pKa = 8.32GPCSGQKK747 pKa = 8.54EE748 pKa = 4.14QPLAEE753 pKa = 4.73LSCSSWWSRR762 pKa = 11.84CEE764 pKa = 3.97CGHH767 pKa = 6.75
Molecular weight: 88.41 kDa
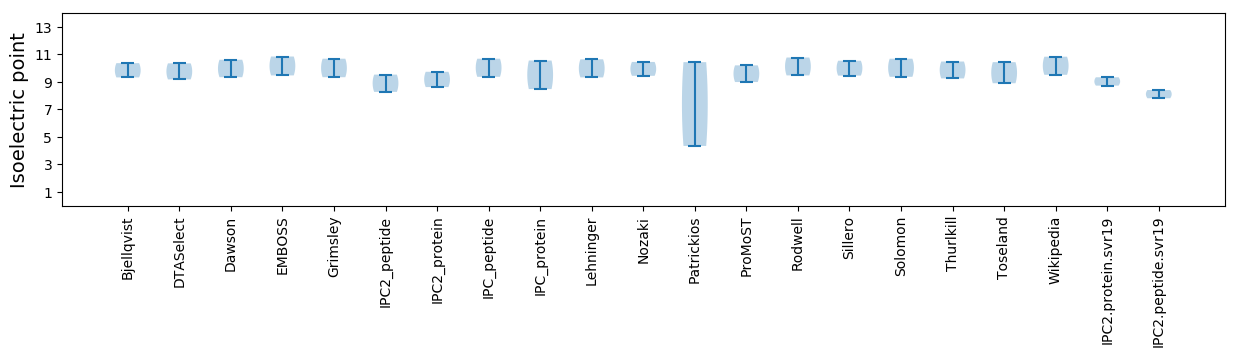
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG98|A0A1L3KG98_9VIRU RNA-directed RNA polymerase OS=Sanxia water strider virus 15 OX=1923399 PE=4 SV=1
MM1 pKa = 7.95DD2 pKa = 5.13LARR5 pKa = 11.84VRR7 pKa = 11.84KK8 pKa = 9.95SNPLLNLAAAAGGAAANVAINKK30 pKa = 8.12VADD33 pKa = 3.86MMRR36 pKa = 11.84PTPPPRR42 pKa = 11.84NARR45 pKa = 11.84RR46 pKa = 11.84QPQMTVAVPRR56 pKa = 11.84NQPRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84KK63 pKa = 9.59QRR65 pKa = 11.84ASNSNNNGLSMARR78 pKa = 11.84QITPGSSIRR87 pKa = 11.84VVDD90 pKa = 3.69TEE92 pKa = 4.26YY93 pKa = 10.54LTVPSTTLNQMQFNPAPPDD112 pKa = 3.49LARR115 pKa = 11.84LEE117 pKa = 4.51QFSKK121 pKa = 9.69MYY123 pKa = 10.46HH124 pKa = 5.38RR125 pKa = 11.84FKK127 pKa = 10.61INRR130 pKa = 11.84ITVHH134 pKa = 5.72YY135 pKa = 9.0EE136 pKa = 3.58PGVGTATTGNISMGICVGPPLGNITTSAHH165 pKa = 5.36IMKK168 pKa = 8.36LTPSCFVPAWKK179 pKa = 10.22SGSITVGRR187 pKa = 11.84DD188 pKa = 2.7IDD190 pKa = 3.11IGRR193 pKa = 11.84YY194 pKa = 6.9MICGGTGDD202 pKa = 3.83TSVACTLYY210 pKa = 10.48TLASAANLGLFRR222 pKa = 11.84VTYY225 pKa = 9.79DD226 pKa = 3.0VSFSYY231 pKa = 10.31PVNFF235 pKa = 4.45
MM1 pKa = 7.95DD2 pKa = 5.13LARR5 pKa = 11.84VRR7 pKa = 11.84KK8 pKa = 9.95SNPLLNLAAAAGGAAANVAINKK30 pKa = 8.12VADD33 pKa = 3.86MMRR36 pKa = 11.84PTPPPRR42 pKa = 11.84NARR45 pKa = 11.84RR46 pKa = 11.84QPQMTVAVPRR56 pKa = 11.84NQPRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84KK63 pKa = 9.59QRR65 pKa = 11.84ASNSNNNGLSMARR78 pKa = 11.84QITPGSSIRR87 pKa = 11.84VVDD90 pKa = 3.69TEE92 pKa = 4.26YY93 pKa = 10.54LTVPSTTLNQMQFNPAPPDD112 pKa = 3.49LARR115 pKa = 11.84LEE117 pKa = 4.51QFSKK121 pKa = 9.69MYY123 pKa = 10.46HH124 pKa = 5.38RR125 pKa = 11.84FKK127 pKa = 10.61INRR130 pKa = 11.84ITVHH134 pKa = 5.72YY135 pKa = 9.0EE136 pKa = 3.58PGVGTATTGNISMGICVGPPLGNITTSAHH165 pKa = 5.36IMKK168 pKa = 8.36LTPSCFVPAWKK179 pKa = 10.22SGSITVGRR187 pKa = 11.84DD188 pKa = 2.7IDD190 pKa = 3.11IGRR193 pKa = 11.84YY194 pKa = 6.9MICGGTGDD202 pKa = 3.83TSVACTLYY210 pKa = 10.48TLASAANLGLFRR222 pKa = 11.84VTYY225 pKa = 9.79DD226 pKa = 3.0VSFSYY231 pKa = 10.31PVNFF235 pKa = 4.45
Molecular weight: 25.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1002 |
235 |
767 |
501.0 |
56.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.886 ± 1.449 | 2.994 ± 0.645 |
3.892 ± 0.244 | 4.491 ± 1.605 |
3.194 ± 0.107 | 5.489 ± 0.871 |
2.794 ± 0.758 | 5.888 ± 0.178 |
5.589 ± 1.303 | 8.184 ± 0.899 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.994 ± 0.63 | 4.691 ± 1.27 |
6.188 ± 0.947 | 3.194 ± 0.105 |
7.685 ± 0.2 | 7.285 ± 0.026 |
6.886 ± 0.811 | 5.489 ± 0.871 |
1.597 ± 0.585 | 4.591 ± 0.805 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
