
Southern bean mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus
Average proteome isoelectric point is 7.17
Get precalculated fractions of proteins
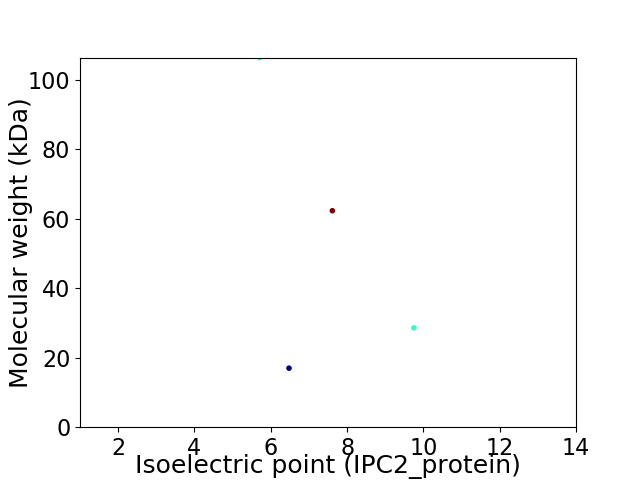
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q7T5V6|Q7T5V6_9VIRU Capsid protein OS=Southern bean mosaic virus OX=12139 PE=3 SV=1
MM1 pKa = 6.78YY2 pKa = 10.23HH3 pKa = 7.46PGRR6 pKa = 11.84SPSFLITLANVICAAILYY24 pKa = 7.55DD25 pKa = 3.43IRR27 pKa = 11.84MGGYY31 pKa = 9.36QPGSLVPIVAWMTPFVTLLWLSASFVTYY59 pKa = 9.74LYY61 pKa = 10.18RR62 pKa = 11.84YY63 pKa = 9.65ARR65 pKa = 11.84TRR67 pKa = 11.84LLPEE71 pKa = 3.85EE72 pKa = 4.09KK73 pKa = 10.13VARR76 pKa = 11.84VYY78 pKa = 9.3YY79 pKa = 8.64TAQSAPYY86 pKa = 8.85FDD88 pKa = 4.77PALGVMMQFAPSHH101 pKa = 6.09GGASIEE107 pKa = 4.27VQVNPSWISLLGGSLKK123 pKa = 10.69INGDD127 pKa = 3.65DD128 pKa = 4.16ASNEE132 pKa = 4.26SAVLGSFYY140 pKa = 11.12SSVKK144 pKa = 10.53PGDD147 pKa = 3.72EE148 pKa = 4.01PASLVAIKK156 pKa = 10.46SGPQTIGFGCRR167 pKa = 11.84TKK169 pKa = 10.47IDD171 pKa = 3.73GDD173 pKa = 3.9DD174 pKa = 4.1CLFTDD179 pKa = 3.15NHH181 pKa = 5.66VWNNSMRR188 pKa = 11.84PTALAKK194 pKa = 10.35AGKK197 pKa = 9.23QVAIEE202 pKa = 4.41DD203 pKa = 3.64WDD205 pKa = 4.24TPLSCDD211 pKa = 3.38HH212 pKa = 6.88KK213 pKa = 10.2MLDD216 pKa = 3.71FVVVRR221 pKa = 11.84VPKK224 pKa = 10.45HH225 pKa = 4.37VWSKK229 pKa = 11.12LGVKK233 pKa = 8.63ATQLVCPSDD242 pKa = 3.81KK243 pKa = 11.17DD244 pKa = 4.1AVTCYY249 pKa = 10.25GGSSSDD255 pKa = 3.6SLLSGTGVCSKK266 pKa = 10.96VDD268 pKa = 3.98FSWKK272 pKa = 8.5LTHH275 pKa = 6.57SCPTAAGWSGTPIYY289 pKa = 10.37SSRR292 pKa = 11.84GVVGMHH298 pKa = 6.33VGFEE302 pKa = 4.97DD303 pKa = 3.14IGKK306 pKa = 9.97LNRR309 pKa = 11.84GVNAFYY315 pKa = 10.97VSNYY319 pKa = 9.68LLRR322 pKa = 11.84SQEE325 pKa = 4.16TLPPDD330 pKa = 3.8LSVIEE335 pKa = 4.5IPFEE339 pKa = 4.26DD340 pKa = 4.14VEE342 pKa = 4.42TRR344 pKa = 11.84SYY346 pKa = 11.47EE347 pKa = 4.17FIEE350 pKa = 4.21VEE352 pKa = 3.54IKK354 pKa = 10.94GRR356 pKa = 11.84GKK358 pKa = 10.49AKK360 pKa = 9.95LGKK363 pKa = 9.86RR364 pKa = 11.84EE365 pKa = 3.99FAWIPEE371 pKa = 4.18SGKK374 pKa = 10.41YY375 pKa = 8.02WADD378 pKa = 3.92DD379 pKa = 4.49DD380 pKa = 5.96DD381 pKa = 6.55DD382 pKa = 5.06SLPPPPKK389 pKa = 10.4VVDD392 pKa = 4.12GKK394 pKa = 9.1MVWTSAQEE402 pKa = 4.18TVAEE406 pKa = 4.27PLNLPEE412 pKa = 4.49GGRR415 pKa = 11.84VKK417 pKa = 10.81ALAALSQLAGYY428 pKa = 8.62NFKK431 pKa = 10.68EE432 pKa = 4.49GEE434 pKa = 4.1AASTRR439 pKa = 11.84GMPLRR444 pKa = 11.84FVGQSACKK452 pKa = 9.92FRR454 pKa = 11.84EE455 pKa = 4.33LCRR458 pKa = 11.84KK459 pKa = 7.66DD460 pKa = 3.48TPDD463 pKa = 2.98EE464 pKa = 3.98VLRR467 pKa = 11.84ATRR470 pKa = 11.84VFPEE474 pKa = 4.61LSDD477 pKa = 3.9FSWPEE482 pKa = 3.65RR483 pKa = 11.84GSKK486 pKa = 10.82AEE488 pKa = 3.92LHH490 pKa = 6.42SLLLQAGKK498 pKa = 10.22FNPTAIPRR506 pKa = 11.84NLEE509 pKa = 3.9GACQNLLEE517 pKa = 5.14RR518 pKa = 11.84YY519 pKa = 7.28PASKK523 pKa = 10.08SCYY526 pKa = 8.82CLRR529 pKa = 11.84GEE531 pKa = 4.39AWSFDD536 pKa = 3.22AVYY539 pKa = 11.09EE540 pKa = 4.32EE541 pKa = 4.65VCKK544 pKa = 10.41KK545 pKa = 10.59AQSAEE550 pKa = 4.12INEE553 pKa = 4.4KK554 pKa = 10.57ASPGVPLSRR563 pKa = 11.84LASTNKK569 pKa = 10.22DD570 pKa = 3.04LLKK573 pKa = 10.72RR574 pKa = 11.84HH575 pKa = 6.36LEE577 pKa = 3.98LVALCVTEE585 pKa = 5.02RR586 pKa = 11.84LFLLSEE592 pKa = 4.9AEE594 pKa = 4.09DD595 pKa = 3.73LHH597 pKa = 6.74NKK599 pKa = 10.05SPVDD603 pKa = 3.8LVQMGLCDD611 pKa = 3.77PVRR614 pKa = 11.84LFVKK618 pKa = 10.09QEE620 pKa = 3.55PHH622 pKa = 6.66ASRR625 pKa = 11.84KK626 pKa = 8.72VKK628 pKa = 10.06EE629 pKa = 3.96GRR631 pKa = 11.84FRR633 pKa = 11.84LISSVSLVDD642 pKa = 3.53QLVEE646 pKa = 4.17RR647 pKa = 11.84MLFGPQNQLEE657 pKa = 4.27IAEE660 pKa = 4.39WEE662 pKa = 4.59HH663 pKa = 7.05IPSKK667 pKa = 10.61PGMGLSLQRR676 pKa = 11.84QAKK679 pKa = 10.16SLFDD683 pKa = 4.03DD684 pKa = 4.2LRR686 pKa = 11.84VKK688 pKa = 10.81HH689 pKa = 5.85SRR691 pKa = 11.84CPAAEE696 pKa = 3.96ADD698 pKa = 3.14ISGFDD703 pKa = 3.43WSVQDD708 pKa = 3.34WEE710 pKa = 4.28LWADD714 pKa = 3.34VEE716 pKa = 4.29MRR718 pKa = 11.84IVLGGFGQKK727 pKa = 10.49LSIAARR733 pKa = 11.84NRR735 pKa = 11.84FSCFMNSVFQLSDD748 pKa = 3.05GTLIEE753 pKa = 4.06QQLPGIMKK761 pKa = 10.12SGSYY765 pKa = 8.61CTSSTNSRR773 pKa = 11.84IRR775 pKa = 11.84CLMAEE780 pKa = 4.85LIGSPWCIAMGDD792 pKa = 3.8DD793 pKa = 5.11SVEE796 pKa = 4.01GWVDD800 pKa = 3.51GAKK803 pKa = 10.31DD804 pKa = 2.83KK805 pKa = 11.33YY806 pKa = 9.83MRR808 pKa = 11.84LGHH811 pKa = 5.36TCKK814 pKa = 10.33DD815 pKa = 3.75YY816 pKa = 11.17KK817 pKa = 10.73PCATSISGRR826 pKa = 11.84LYY828 pKa = 9.38EE829 pKa = 5.04VEE831 pKa = 4.31FCSHH835 pKa = 7.24VIRR838 pKa = 11.84EE839 pKa = 4.21DD840 pKa = 3.82RR841 pKa = 11.84CWLASWPKK849 pKa = 9.39TLYY852 pKa = 10.46KK853 pKa = 10.46YY854 pKa = 10.66LSEE857 pKa = 4.23GKK859 pKa = 9.12WFFEE863 pKa = 4.14DD864 pKa = 4.39LEE866 pKa = 5.46RR867 pKa = 11.84EE868 pKa = 4.45LGSSPHH874 pKa = 6.03WPRR877 pKa = 11.84IRR879 pKa = 11.84HH880 pKa = 4.43YY881 pKa = 11.18VVGNTPSPDD890 pKa = 2.95KK891 pKa = 10.92TRR893 pKa = 11.84LEE895 pKa = 4.23NSSPSYY901 pKa = 11.07GEE903 pKa = 4.2EE904 pKa = 3.8ADD906 pKa = 3.77KK907 pKa = 8.87TTVSQGYY914 pKa = 8.23SEE916 pKa = 5.34HH917 pKa = 6.7SGSPGHH923 pKa = 6.54SIEE926 pKa = 4.28EE927 pKa = 4.08AQEE930 pKa = 4.07PEE932 pKa = 4.2TAPFCCKK939 pKa = 10.07AASVYY944 pKa = 10.02PGWGIHH950 pKa = 6.47GPYY953 pKa = 10.04CSGGYY958 pKa = 10.45GSLTT962 pKa = 3.36
MM1 pKa = 6.78YY2 pKa = 10.23HH3 pKa = 7.46PGRR6 pKa = 11.84SPSFLITLANVICAAILYY24 pKa = 7.55DD25 pKa = 3.43IRR27 pKa = 11.84MGGYY31 pKa = 9.36QPGSLVPIVAWMTPFVTLLWLSASFVTYY59 pKa = 9.74LYY61 pKa = 10.18RR62 pKa = 11.84YY63 pKa = 9.65ARR65 pKa = 11.84TRR67 pKa = 11.84LLPEE71 pKa = 3.85EE72 pKa = 4.09KK73 pKa = 10.13VARR76 pKa = 11.84VYY78 pKa = 9.3YY79 pKa = 8.64TAQSAPYY86 pKa = 8.85FDD88 pKa = 4.77PALGVMMQFAPSHH101 pKa = 6.09GGASIEE107 pKa = 4.27VQVNPSWISLLGGSLKK123 pKa = 10.69INGDD127 pKa = 3.65DD128 pKa = 4.16ASNEE132 pKa = 4.26SAVLGSFYY140 pKa = 11.12SSVKK144 pKa = 10.53PGDD147 pKa = 3.72EE148 pKa = 4.01PASLVAIKK156 pKa = 10.46SGPQTIGFGCRR167 pKa = 11.84TKK169 pKa = 10.47IDD171 pKa = 3.73GDD173 pKa = 3.9DD174 pKa = 4.1CLFTDD179 pKa = 3.15NHH181 pKa = 5.66VWNNSMRR188 pKa = 11.84PTALAKK194 pKa = 10.35AGKK197 pKa = 9.23QVAIEE202 pKa = 4.41DD203 pKa = 3.64WDD205 pKa = 4.24TPLSCDD211 pKa = 3.38HH212 pKa = 6.88KK213 pKa = 10.2MLDD216 pKa = 3.71FVVVRR221 pKa = 11.84VPKK224 pKa = 10.45HH225 pKa = 4.37VWSKK229 pKa = 11.12LGVKK233 pKa = 8.63ATQLVCPSDD242 pKa = 3.81KK243 pKa = 11.17DD244 pKa = 4.1AVTCYY249 pKa = 10.25GGSSSDD255 pKa = 3.6SLLSGTGVCSKK266 pKa = 10.96VDD268 pKa = 3.98FSWKK272 pKa = 8.5LTHH275 pKa = 6.57SCPTAAGWSGTPIYY289 pKa = 10.37SSRR292 pKa = 11.84GVVGMHH298 pKa = 6.33VGFEE302 pKa = 4.97DD303 pKa = 3.14IGKK306 pKa = 9.97LNRR309 pKa = 11.84GVNAFYY315 pKa = 10.97VSNYY319 pKa = 9.68LLRR322 pKa = 11.84SQEE325 pKa = 4.16TLPPDD330 pKa = 3.8LSVIEE335 pKa = 4.5IPFEE339 pKa = 4.26DD340 pKa = 4.14VEE342 pKa = 4.42TRR344 pKa = 11.84SYY346 pKa = 11.47EE347 pKa = 4.17FIEE350 pKa = 4.21VEE352 pKa = 3.54IKK354 pKa = 10.94GRR356 pKa = 11.84GKK358 pKa = 10.49AKK360 pKa = 9.95LGKK363 pKa = 9.86RR364 pKa = 11.84EE365 pKa = 3.99FAWIPEE371 pKa = 4.18SGKK374 pKa = 10.41YY375 pKa = 8.02WADD378 pKa = 3.92DD379 pKa = 4.49DD380 pKa = 5.96DD381 pKa = 6.55DD382 pKa = 5.06SLPPPPKK389 pKa = 10.4VVDD392 pKa = 4.12GKK394 pKa = 9.1MVWTSAQEE402 pKa = 4.18TVAEE406 pKa = 4.27PLNLPEE412 pKa = 4.49GGRR415 pKa = 11.84VKK417 pKa = 10.81ALAALSQLAGYY428 pKa = 8.62NFKK431 pKa = 10.68EE432 pKa = 4.49GEE434 pKa = 4.1AASTRR439 pKa = 11.84GMPLRR444 pKa = 11.84FVGQSACKK452 pKa = 9.92FRR454 pKa = 11.84EE455 pKa = 4.33LCRR458 pKa = 11.84KK459 pKa = 7.66DD460 pKa = 3.48TPDD463 pKa = 2.98EE464 pKa = 3.98VLRR467 pKa = 11.84ATRR470 pKa = 11.84VFPEE474 pKa = 4.61LSDD477 pKa = 3.9FSWPEE482 pKa = 3.65RR483 pKa = 11.84GSKK486 pKa = 10.82AEE488 pKa = 3.92LHH490 pKa = 6.42SLLLQAGKK498 pKa = 10.22FNPTAIPRR506 pKa = 11.84NLEE509 pKa = 3.9GACQNLLEE517 pKa = 5.14RR518 pKa = 11.84YY519 pKa = 7.28PASKK523 pKa = 10.08SCYY526 pKa = 8.82CLRR529 pKa = 11.84GEE531 pKa = 4.39AWSFDD536 pKa = 3.22AVYY539 pKa = 11.09EE540 pKa = 4.32EE541 pKa = 4.65VCKK544 pKa = 10.41KK545 pKa = 10.59AQSAEE550 pKa = 4.12INEE553 pKa = 4.4KK554 pKa = 10.57ASPGVPLSRR563 pKa = 11.84LASTNKK569 pKa = 10.22DD570 pKa = 3.04LLKK573 pKa = 10.72RR574 pKa = 11.84HH575 pKa = 6.36LEE577 pKa = 3.98LVALCVTEE585 pKa = 5.02RR586 pKa = 11.84LFLLSEE592 pKa = 4.9AEE594 pKa = 4.09DD595 pKa = 3.73LHH597 pKa = 6.74NKK599 pKa = 10.05SPVDD603 pKa = 3.8LVQMGLCDD611 pKa = 3.77PVRR614 pKa = 11.84LFVKK618 pKa = 10.09QEE620 pKa = 3.55PHH622 pKa = 6.66ASRR625 pKa = 11.84KK626 pKa = 8.72VKK628 pKa = 10.06EE629 pKa = 3.96GRR631 pKa = 11.84FRR633 pKa = 11.84LISSVSLVDD642 pKa = 3.53QLVEE646 pKa = 4.17RR647 pKa = 11.84MLFGPQNQLEE657 pKa = 4.27IAEE660 pKa = 4.39WEE662 pKa = 4.59HH663 pKa = 7.05IPSKK667 pKa = 10.61PGMGLSLQRR676 pKa = 11.84QAKK679 pKa = 10.16SLFDD683 pKa = 4.03DD684 pKa = 4.2LRR686 pKa = 11.84VKK688 pKa = 10.81HH689 pKa = 5.85SRR691 pKa = 11.84CPAAEE696 pKa = 3.96ADD698 pKa = 3.14ISGFDD703 pKa = 3.43WSVQDD708 pKa = 3.34WEE710 pKa = 4.28LWADD714 pKa = 3.34VEE716 pKa = 4.29MRR718 pKa = 11.84IVLGGFGQKK727 pKa = 10.49LSIAARR733 pKa = 11.84NRR735 pKa = 11.84FSCFMNSVFQLSDD748 pKa = 3.05GTLIEE753 pKa = 4.06QQLPGIMKK761 pKa = 10.12SGSYY765 pKa = 8.61CTSSTNSRR773 pKa = 11.84IRR775 pKa = 11.84CLMAEE780 pKa = 4.85LIGSPWCIAMGDD792 pKa = 3.8DD793 pKa = 5.11SVEE796 pKa = 4.01GWVDD800 pKa = 3.51GAKK803 pKa = 10.31DD804 pKa = 2.83KK805 pKa = 11.33YY806 pKa = 9.83MRR808 pKa = 11.84LGHH811 pKa = 5.36TCKK814 pKa = 10.33DD815 pKa = 3.75YY816 pKa = 11.17KK817 pKa = 10.73PCATSISGRR826 pKa = 11.84LYY828 pKa = 9.38EE829 pKa = 5.04VEE831 pKa = 4.31FCSHH835 pKa = 7.24VIRR838 pKa = 11.84EE839 pKa = 4.21DD840 pKa = 3.82RR841 pKa = 11.84CWLASWPKK849 pKa = 9.39TLYY852 pKa = 10.46KK853 pKa = 10.46YY854 pKa = 10.66LSEE857 pKa = 4.23GKK859 pKa = 9.12WFFEE863 pKa = 4.14DD864 pKa = 4.39LEE866 pKa = 5.46RR867 pKa = 11.84EE868 pKa = 4.45LGSSPHH874 pKa = 6.03WPRR877 pKa = 11.84IRR879 pKa = 11.84HH880 pKa = 4.43YY881 pKa = 11.18VVGNTPSPDD890 pKa = 2.95KK891 pKa = 10.92TRR893 pKa = 11.84LEE895 pKa = 4.23NSSPSYY901 pKa = 11.07GEE903 pKa = 4.2EE904 pKa = 3.8ADD906 pKa = 3.77KK907 pKa = 8.87TTVSQGYY914 pKa = 8.23SEE916 pKa = 5.34HH917 pKa = 6.7SGSPGHH923 pKa = 6.54SIEE926 pKa = 4.28EE927 pKa = 4.08AQEE930 pKa = 4.07PEE932 pKa = 4.2TAPFCCKK939 pKa = 10.07AASVYY944 pKa = 10.02PGWGIHH950 pKa = 6.47GPYY953 pKa = 10.04CSGGYY958 pKa = 10.45GSLTT962 pKa = 3.36
Molecular weight: 106.44 kDa
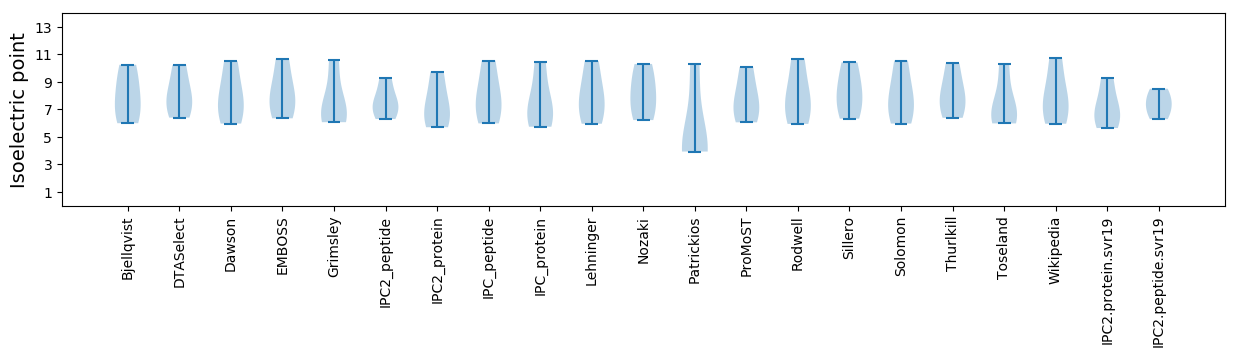
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q7T5V7|Q7T5V7_9VIRU Putative movement protein OS=Southern bean mosaic virus OX=12139 PE=4 SV=1
MM1 pKa = 7.87AKK3 pKa = 10.33RR4 pKa = 11.84LTKK7 pKa = 10.22QQLAKK12 pKa = 10.68AIANTLEE19 pKa = 4.14APATQSRR26 pKa = 11.84RR27 pKa = 11.84PRR29 pKa = 11.84NRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SAARR39 pKa = 11.84QPQSIQAGASMAPIAQGAMVRR60 pKa = 11.84LRR62 pKa = 11.84EE63 pKa = 4.0PSLRR67 pKa = 11.84TAGGVTVLTHH77 pKa = 6.43SEE79 pKa = 4.22LSTEE83 pKa = 4.12LAVTNAIVVSSEE95 pKa = 3.45LVMPFTMGTWLRR107 pKa = 11.84GVAANWSKK115 pKa = 11.55YY116 pKa = 7.81SLFSVRR122 pKa = 11.84YY123 pKa = 7.48TYY125 pKa = 10.98LPSCPSTTSGSIHH138 pKa = 6.61MGFQYY143 pKa = 11.56DD144 pKa = 3.61MADD147 pKa = 3.6TLPVSVNQLSNLRR160 pKa = 11.84GYY162 pKa = 11.09VSGQVWSGSSGLCYY176 pKa = 10.29INGTRR181 pKa = 11.84CSDD184 pKa = 3.37TANAITTTLDD194 pKa = 3.15VAKK197 pKa = 10.16LGKK200 pKa = 9.39KK201 pKa = 8.78WYY203 pKa = 8.93PFKK206 pKa = 10.78TSTDD210 pKa = 3.48FTAAVGVIVNIATPLVPARR229 pKa = 11.84LVIAMLDD236 pKa = 3.92GSSSTAVSTGRR247 pKa = 11.84LYY249 pKa = 11.25VSYY252 pKa = 9.06TVQLIEE258 pKa = 4.15PTALALNNN266 pKa = 3.79
MM1 pKa = 7.87AKK3 pKa = 10.33RR4 pKa = 11.84LTKK7 pKa = 10.22QQLAKK12 pKa = 10.68AIANTLEE19 pKa = 4.14APATQSRR26 pKa = 11.84RR27 pKa = 11.84PRR29 pKa = 11.84NRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SAARR39 pKa = 11.84QPQSIQAGASMAPIAQGAMVRR60 pKa = 11.84LRR62 pKa = 11.84EE63 pKa = 4.0PSLRR67 pKa = 11.84TAGGVTVLTHH77 pKa = 6.43SEE79 pKa = 4.22LSTEE83 pKa = 4.12LAVTNAIVVSSEE95 pKa = 3.45LVMPFTMGTWLRR107 pKa = 11.84GVAANWSKK115 pKa = 11.55YY116 pKa = 7.81SLFSVRR122 pKa = 11.84YY123 pKa = 7.48TYY125 pKa = 10.98LPSCPSTTSGSIHH138 pKa = 6.61MGFQYY143 pKa = 11.56DD144 pKa = 3.61MADD147 pKa = 3.6TLPVSVNQLSNLRR160 pKa = 11.84GYY162 pKa = 11.09VSGQVWSGSSGLCYY176 pKa = 10.29INGTRR181 pKa = 11.84CSDD184 pKa = 3.37TANAITTTLDD194 pKa = 3.15VAKK197 pKa = 10.16LGKK200 pKa = 9.39KK201 pKa = 8.78WYY203 pKa = 8.93PFKK206 pKa = 10.78TSTDD210 pKa = 3.48FTAAVGVIVNIATPLVPARR229 pKa = 11.84LVIAMLDD236 pKa = 3.92GSSSTAVSTGRR247 pKa = 11.84LYY249 pKa = 11.25VSYY252 pKa = 9.06TVQLIEE258 pKa = 4.15PTALALNNN266 pKa = 3.79
Molecular weight: 28.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1946 |
147 |
962 |
486.5 |
53.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.4 ± 1.052 | 2.518 ± 0.461 |
4.573 ± 0.692 | 5.653 ± 0.951 |
3.649 ± 0.9 | 7.297 ± 0.438 |
1.85 ± 0.609 | 4.111 ± 0.524 |
5.293 ± 0.776 | 8.941 ± 0.544 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.953 ± 0.325 | 2.826 ± 0.372 |
6.526 ± 1.038 | 3.34 ± 0.293 |
5.396 ± 0.683 | 10.175 ± 0.823 |
5.55 ± 1.284 | 7.554 ± 0.608 |
2.107 ± 0.283 | 3.289 ± 0.057 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
