
Methylopila sp. Yamaguchi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylocystaceae; Methylopila; unclassified Methylopila
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
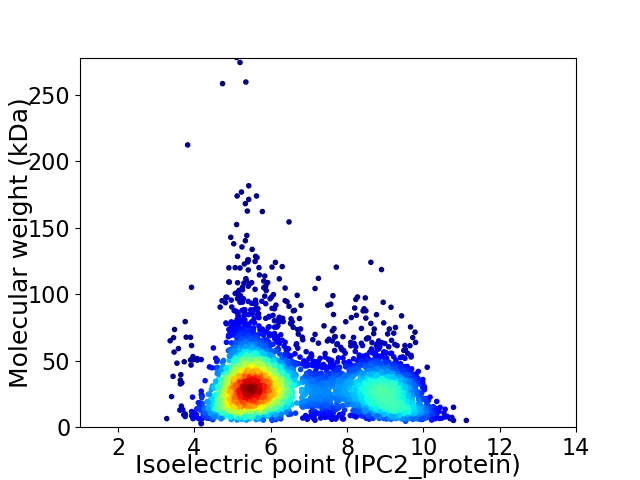
Virtual 2D-PAGE plot for 4120 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H6BDX8|A0A2H6BDX8_9RHIZ NUDIX hydrolase OS=Methylopila sp. Yamaguchi OX=1437817 GN=METY_1595 PE=3 SV=1
MM1 pKa = 6.92ITINAKK7 pKa = 10.24SAVFDD12 pKa = 3.3ATGIDD17 pKa = 3.51FEE19 pKa = 5.21AYY21 pKa = 10.11VRR23 pKa = 11.84GQFVADD29 pKa = 3.5ATGSGFPVFDD39 pKa = 3.76NSSAFSGEE47 pKa = 3.86EE48 pKa = 3.52MFIGYY53 pKa = 7.89GTEE56 pKa = 3.89ATSKK60 pKa = 10.48YY61 pKa = 10.22VLAHH65 pKa = 5.58GQVEE69 pKa = 4.79YY70 pKa = 11.25AFGTHH75 pKa = 5.33TVAGTINTIEE85 pKa = 4.24YY86 pKa = 8.19GTIGEE91 pKa = 4.44GSYY94 pKa = 10.8DD95 pKa = 3.47ANGYY99 pKa = 7.32FTGGNVALTITGLDD113 pKa = 3.52LSNPVPADD121 pKa = 3.15NAEE124 pKa = 4.09AVGIEE129 pKa = 4.11ATGAVHH135 pKa = 6.82NFALAHH141 pKa = 5.99MYY143 pKa = 10.49GSSGDD148 pKa = 3.39QARR151 pKa = 11.84FDD153 pKa = 4.11TYY155 pKa = 11.81ADD157 pKa = 3.83ALDD160 pKa = 4.82SEE162 pKa = 4.82AQTFLGSRR170 pKa = 11.84GNDD173 pKa = 3.22VYY175 pKa = 11.61VGTAFNDD182 pKa = 3.93VIRR185 pKa = 11.84SNAGDD190 pKa = 3.65DD191 pKa = 4.03RR192 pKa = 11.84ITGGGGDD199 pKa = 3.7DD200 pKa = 4.28KK201 pKa = 11.17IYY203 pKa = 10.78GSRR206 pKa = 11.84GVDD209 pKa = 3.12TAVYY213 pKa = 10.11SGDD216 pKa = 3.46RR217 pKa = 11.84ADD219 pKa = 3.77YY220 pKa = 10.94AITRR224 pKa = 11.84DD225 pKa = 3.59LAGVLHH231 pKa = 6.24VVDD234 pKa = 5.6LRR236 pKa = 11.84AGATSDD242 pKa = 4.53GADD245 pKa = 2.93QLVDD249 pKa = 3.54VEE251 pKa = 4.37QLEE254 pKa = 4.56FADD257 pKa = 4.28GTLDD261 pKa = 3.55ASTIAAGGMLTLDD274 pKa = 3.62ASGASTGVDD283 pKa = 3.12MTTFLADD290 pKa = 3.9FFAGVATGAAYY301 pKa = 9.56KK302 pKa = 10.3FYY304 pKa = 11.08GGTPDD309 pKa = 3.15QAFGGTYY316 pKa = 10.46YY317 pKa = 10.73MNGDD321 pKa = 3.41QLAYY325 pKa = 10.13QYY327 pKa = 10.89TEE329 pKa = 4.27GGVATDD335 pKa = 3.15ARR337 pKa = 11.84LVFGGEE343 pKa = 3.59EE344 pKa = 3.49LAYY347 pKa = 10.6DD348 pKa = 5.12FIHH351 pKa = 6.97HH352 pKa = 6.79GSQYY356 pKa = 10.54GHH358 pKa = 7.52GITGALDD365 pKa = 3.48SLTFGAWTADD375 pKa = 3.41TTGTEE380 pKa = 4.66GVGSAGLIQNLYY392 pKa = 9.49EE393 pKa = 4.08ALKK396 pKa = 9.29ITGLGFDD403 pKa = 3.91VAPGAGSSNPVHH415 pKa = 6.8LLHH418 pKa = 6.81AAARR422 pKa = 11.84AGDD425 pKa = 3.51ASVFEE430 pKa = 5.18DD431 pKa = 4.29LLASRR436 pKa = 11.84PQHH439 pKa = 6.57FIGSDD444 pKa = 3.36GDD446 pKa = 3.81DD447 pKa = 3.74VYY449 pKa = 11.64VGSAFADD456 pKa = 3.87VVEE459 pKa = 5.22GGAGADD465 pKa = 3.89TLDD468 pKa = 4.11GGQGADD474 pKa = 3.56TLSGGLGDD482 pKa = 3.71DD483 pKa = 3.83TYY485 pKa = 11.92YY486 pKa = 11.15VDD488 pKa = 3.59NSRR491 pKa = 11.84DD492 pKa = 3.73TVIEE496 pKa = 3.93AAGEE500 pKa = 4.14GADD503 pKa = 4.15TIVASADD510 pKa = 3.47FKK512 pKa = 11.28LGARR516 pKa = 11.84VAIEE520 pKa = 3.78TLIAADD526 pKa = 4.8GSDD529 pKa = 4.3ADD531 pKa = 4.07LTANRR536 pKa = 11.84YY537 pKa = 7.7VRR539 pKa = 11.84NLIGSDD545 pKa = 2.92GDD547 pKa = 3.57NRR549 pKa = 11.84LDD551 pKa = 3.45AFAGAKK557 pKa = 8.61TVTGGAGEE565 pKa = 4.21DD566 pKa = 3.53TFVFTVDD573 pKa = 3.16VRR575 pKa = 11.84KK576 pKa = 9.73SAAVISDD583 pKa = 3.76FTSGEE588 pKa = 3.93DD589 pKa = 4.25SIEE592 pKa = 4.11LSSKK596 pKa = 10.21VFTALGVGALDD607 pKa = 3.75AGSFVLGSAALDD619 pKa = 3.89ADD621 pKa = 4.01DD622 pKa = 6.06HH623 pKa = 7.14ILYY626 pKa = 10.06DD627 pKa = 3.96ASTGSLFYY635 pKa = 11.01DD636 pKa = 3.6ADD638 pKa = 4.09GVGGSAALLFATLSSAPSLSAADD661 pKa = 3.83FLVAA665 pKa = 5.42
MM1 pKa = 6.92ITINAKK7 pKa = 10.24SAVFDD12 pKa = 3.3ATGIDD17 pKa = 3.51FEE19 pKa = 5.21AYY21 pKa = 10.11VRR23 pKa = 11.84GQFVADD29 pKa = 3.5ATGSGFPVFDD39 pKa = 3.76NSSAFSGEE47 pKa = 3.86EE48 pKa = 3.52MFIGYY53 pKa = 7.89GTEE56 pKa = 3.89ATSKK60 pKa = 10.48YY61 pKa = 10.22VLAHH65 pKa = 5.58GQVEE69 pKa = 4.79YY70 pKa = 11.25AFGTHH75 pKa = 5.33TVAGTINTIEE85 pKa = 4.24YY86 pKa = 8.19GTIGEE91 pKa = 4.44GSYY94 pKa = 10.8DD95 pKa = 3.47ANGYY99 pKa = 7.32FTGGNVALTITGLDD113 pKa = 3.52LSNPVPADD121 pKa = 3.15NAEE124 pKa = 4.09AVGIEE129 pKa = 4.11ATGAVHH135 pKa = 6.82NFALAHH141 pKa = 5.99MYY143 pKa = 10.49GSSGDD148 pKa = 3.39QARR151 pKa = 11.84FDD153 pKa = 4.11TYY155 pKa = 11.81ADD157 pKa = 3.83ALDD160 pKa = 4.82SEE162 pKa = 4.82AQTFLGSRR170 pKa = 11.84GNDD173 pKa = 3.22VYY175 pKa = 11.61VGTAFNDD182 pKa = 3.93VIRR185 pKa = 11.84SNAGDD190 pKa = 3.65DD191 pKa = 4.03RR192 pKa = 11.84ITGGGGDD199 pKa = 3.7DD200 pKa = 4.28KK201 pKa = 11.17IYY203 pKa = 10.78GSRR206 pKa = 11.84GVDD209 pKa = 3.12TAVYY213 pKa = 10.11SGDD216 pKa = 3.46RR217 pKa = 11.84ADD219 pKa = 3.77YY220 pKa = 10.94AITRR224 pKa = 11.84DD225 pKa = 3.59LAGVLHH231 pKa = 6.24VVDD234 pKa = 5.6LRR236 pKa = 11.84AGATSDD242 pKa = 4.53GADD245 pKa = 2.93QLVDD249 pKa = 3.54VEE251 pKa = 4.37QLEE254 pKa = 4.56FADD257 pKa = 4.28GTLDD261 pKa = 3.55ASTIAAGGMLTLDD274 pKa = 3.62ASGASTGVDD283 pKa = 3.12MTTFLADD290 pKa = 3.9FFAGVATGAAYY301 pKa = 9.56KK302 pKa = 10.3FYY304 pKa = 11.08GGTPDD309 pKa = 3.15QAFGGTYY316 pKa = 10.46YY317 pKa = 10.73MNGDD321 pKa = 3.41QLAYY325 pKa = 10.13QYY327 pKa = 10.89TEE329 pKa = 4.27GGVATDD335 pKa = 3.15ARR337 pKa = 11.84LVFGGEE343 pKa = 3.59EE344 pKa = 3.49LAYY347 pKa = 10.6DD348 pKa = 5.12FIHH351 pKa = 6.97HH352 pKa = 6.79GSQYY356 pKa = 10.54GHH358 pKa = 7.52GITGALDD365 pKa = 3.48SLTFGAWTADD375 pKa = 3.41TTGTEE380 pKa = 4.66GVGSAGLIQNLYY392 pKa = 9.49EE393 pKa = 4.08ALKK396 pKa = 9.29ITGLGFDD403 pKa = 3.91VAPGAGSSNPVHH415 pKa = 6.8LLHH418 pKa = 6.81AAARR422 pKa = 11.84AGDD425 pKa = 3.51ASVFEE430 pKa = 5.18DD431 pKa = 4.29LLASRR436 pKa = 11.84PQHH439 pKa = 6.57FIGSDD444 pKa = 3.36GDD446 pKa = 3.81DD447 pKa = 3.74VYY449 pKa = 11.64VGSAFADD456 pKa = 3.87VVEE459 pKa = 5.22GGAGADD465 pKa = 3.89TLDD468 pKa = 4.11GGQGADD474 pKa = 3.56TLSGGLGDD482 pKa = 3.71DD483 pKa = 3.83TYY485 pKa = 11.92YY486 pKa = 11.15VDD488 pKa = 3.59NSRR491 pKa = 11.84DD492 pKa = 3.73TVIEE496 pKa = 3.93AAGEE500 pKa = 4.14GADD503 pKa = 4.15TIVASADD510 pKa = 3.47FKK512 pKa = 11.28LGARR516 pKa = 11.84VAIEE520 pKa = 3.78TLIAADD526 pKa = 4.8GSDD529 pKa = 4.3ADD531 pKa = 4.07LTANRR536 pKa = 11.84YY537 pKa = 7.7VRR539 pKa = 11.84NLIGSDD545 pKa = 2.92GDD547 pKa = 3.57NRR549 pKa = 11.84LDD551 pKa = 3.45AFAGAKK557 pKa = 8.61TVTGGAGEE565 pKa = 4.21DD566 pKa = 3.53TFVFTVDD573 pKa = 3.16VRR575 pKa = 11.84KK576 pKa = 9.73SAAVISDD583 pKa = 3.76FTSGEE588 pKa = 3.93DD589 pKa = 4.25SIEE592 pKa = 4.11LSSKK596 pKa = 10.21VFTALGVGALDD607 pKa = 3.75AGSFVLGSAALDD619 pKa = 3.89ADD621 pKa = 4.01DD622 pKa = 6.06HH623 pKa = 7.14ILYY626 pKa = 10.06DD627 pKa = 3.96ASTGSLFYY635 pKa = 11.01DD636 pKa = 3.6ADD638 pKa = 4.09GVGGSAALLFATLSSAPSLSAADD661 pKa = 3.83FLVAA665 pKa = 5.42
Molecular weight: 67.76 kDa
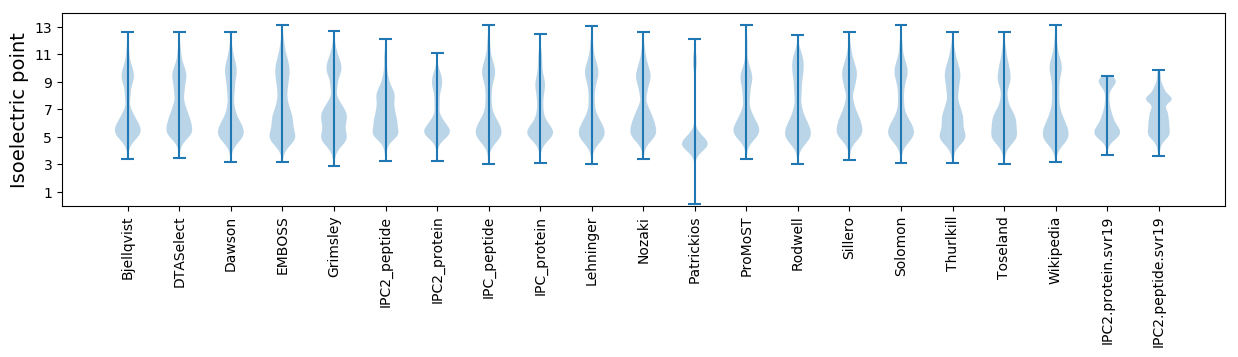
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H6BJM6|A0A2H6BJM6_9RHIZ Molybdopterin dehydrogenase FAD-binding OS=Methylopila sp. Yamaguchi OX=1437817 GN=METY_3600 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.31VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.89GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.31VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.89GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1302561 |
26 |
2604 |
316.2 |
33.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.658 ± 0.064 | 0.714 ± 0.011 |
5.817 ± 0.035 | 5.695 ± 0.038 |
3.601 ± 0.025 | 8.976 ± 0.035 |
1.786 ± 0.019 | 4.317 ± 0.028 |
2.975 ± 0.03 | 10.212 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.092 ± 0.016 | 2.092 ± 0.021 |
5.467 ± 0.029 | 2.519 ± 0.023 |
7.706 ± 0.038 | 4.962 ± 0.022 |
5.113 ± 0.027 | 8.043 ± 0.035 |
1.259 ± 0.015 | 1.998 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
