
Pelolinea submarina
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Anaerolineae; Anaerolineales; Anaerolineaceae; Pelolinea
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
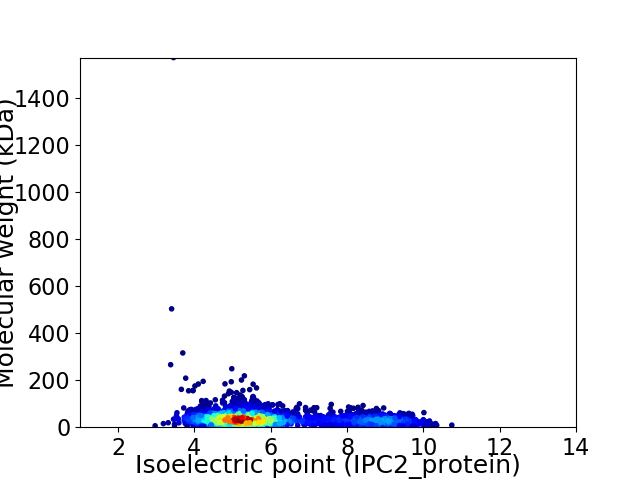
Virtual 2D-PAGE plot for 3066 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A347ZPX9|A0A347ZPX9_9CHLR PTS system IIB component (L-Asc family) OS=Pelolinea submarina OX=913107 GN=DFR64_2744 PE=4 SV=1
MM1 pKa = 7.56ISTRR5 pKa = 11.84SVRR8 pKa = 11.84LISVLFLATMLLTACGGSSTAAASDD33 pKa = 3.37WSNAASAEE41 pKa = 4.17AGGGMDD47 pKa = 4.55ALVAAAQAEE56 pKa = 4.63GEE58 pKa = 4.5LNVIALPEE66 pKa = 4.2DD67 pKa = 3.02WCNYY71 pKa = 9.42GGMIATFEE79 pKa = 4.28DD80 pKa = 4.11KK81 pKa = 11.3YY82 pKa = 10.84GIKK85 pKa = 10.4VNSITPEE92 pKa = 4.01AGSADD97 pKa = 3.94EE98 pKa = 4.2VQAIIDD104 pKa = 3.66NKK106 pKa = 9.46EE107 pKa = 3.97NKK109 pKa = 9.71GPQAPDD115 pKa = 3.49VIDD118 pKa = 3.4VGPAYY123 pKa = 10.28GPSSKK128 pKa = 10.92DD129 pKa = 3.18QDD131 pKa = 3.8LLAPYY136 pKa = 10.14KK137 pKa = 9.63VTTWDD142 pKa = 3.02SMTGVKK148 pKa = 10.37DD149 pKa = 3.26ADD151 pKa = 3.55GYY153 pKa = 11.78YY154 pKa = 8.04YY155 pKa = 10.56TDD157 pKa = 3.44YY158 pKa = 10.97NGVMVFEE165 pKa = 4.81VNTDD169 pKa = 3.06VVTDD173 pKa = 3.88VPQDD177 pKa = 3.7YY178 pKa = 11.51ADD180 pKa = 4.84LLDD183 pKa = 4.15PKK185 pKa = 11.33YY186 pKa = 10.64NGQVALAGDD195 pKa = 3.99PRR197 pKa = 11.84ASNQAAQTVYY207 pKa = 10.9AAALANGGSLDD218 pKa = 4.63DD219 pKa = 3.86IQPGLEE225 pKa = 3.97YY226 pKa = 10.64FKK228 pKa = 11.28ALNEE232 pKa = 4.26NGNLLPLIANTGSIAMGEE250 pKa = 4.29TPITFQWNYY259 pKa = 10.21LALANDD265 pKa = 4.29DD266 pKa = 4.55AFAGNPPLEE275 pKa = 4.12IVYY278 pKa = 7.33PTSVSWGGYY287 pKa = 6.89YY288 pKa = 10.33LQAISAYY295 pKa = 9.54APHH298 pKa = 7.24PAAARR303 pKa = 11.84LWQEE307 pKa = 3.52FLYY310 pKa = 10.77SDD312 pKa = 4.16EE313 pKa = 4.76GQTIWMTGYY322 pKa = 10.04CAPARR327 pKa = 11.84LADD330 pKa = 3.55MLEE333 pKa = 4.31RR334 pKa = 11.84GVVSADD340 pKa = 3.61LQTKK344 pKa = 10.36LPDD347 pKa = 3.65PSIIANAVVPDD358 pKa = 4.17GDD360 pKa = 3.74QLSAARR366 pKa = 11.84DD367 pKa = 3.84LIKK370 pKa = 10.51EE371 pKa = 3.93QWDD374 pKa = 3.98SVVGLDD380 pKa = 4.59IKK382 pKa = 10.73EE383 pKa = 3.96
MM1 pKa = 7.56ISTRR5 pKa = 11.84SVRR8 pKa = 11.84LISVLFLATMLLTACGGSSTAAASDD33 pKa = 3.37WSNAASAEE41 pKa = 4.17AGGGMDD47 pKa = 4.55ALVAAAQAEE56 pKa = 4.63GEE58 pKa = 4.5LNVIALPEE66 pKa = 4.2DD67 pKa = 3.02WCNYY71 pKa = 9.42GGMIATFEE79 pKa = 4.28DD80 pKa = 4.11KK81 pKa = 11.3YY82 pKa = 10.84GIKK85 pKa = 10.4VNSITPEE92 pKa = 4.01AGSADD97 pKa = 3.94EE98 pKa = 4.2VQAIIDD104 pKa = 3.66NKK106 pKa = 9.46EE107 pKa = 3.97NKK109 pKa = 9.71GPQAPDD115 pKa = 3.49VIDD118 pKa = 3.4VGPAYY123 pKa = 10.28GPSSKK128 pKa = 10.92DD129 pKa = 3.18QDD131 pKa = 3.8LLAPYY136 pKa = 10.14KK137 pKa = 9.63VTTWDD142 pKa = 3.02SMTGVKK148 pKa = 10.37DD149 pKa = 3.26ADD151 pKa = 3.55GYY153 pKa = 11.78YY154 pKa = 8.04YY155 pKa = 10.56TDD157 pKa = 3.44YY158 pKa = 10.97NGVMVFEE165 pKa = 4.81VNTDD169 pKa = 3.06VVTDD173 pKa = 3.88VPQDD177 pKa = 3.7YY178 pKa = 11.51ADD180 pKa = 4.84LLDD183 pKa = 4.15PKK185 pKa = 11.33YY186 pKa = 10.64NGQVALAGDD195 pKa = 3.99PRR197 pKa = 11.84ASNQAAQTVYY207 pKa = 10.9AAALANGGSLDD218 pKa = 4.63DD219 pKa = 3.86IQPGLEE225 pKa = 3.97YY226 pKa = 10.64FKK228 pKa = 11.28ALNEE232 pKa = 4.26NGNLLPLIANTGSIAMGEE250 pKa = 4.29TPITFQWNYY259 pKa = 10.21LALANDD265 pKa = 4.29DD266 pKa = 4.55AFAGNPPLEE275 pKa = 4.12IVYY278 pKa = 7.33PTSVSWGGYY287 pKa = 6.89YY288 pKa = 10.33LQAISAYY295 pKa = 9.54APHH298 pKa = 7.24PAAARR303 pKa = 11.84LWQEE307 pKa = 3.52FLYY310 pKa = 10.77SDD312 pKa = 4.16EE313 pKa = 4.76GQTIWMTGYY322 pKa = 10.04CAPARR327 pKa = 11.84LADD330 pKa = 3.55MLEE333 pKa = 4.31RR334 pKa = 11.84GVVSADD340 pKa = 3.61LQTKK344 pKa = 10.36LPDD347 pKa = 3.65PSIIANAVVPDD358 pKa = 4.17GDD360 pKa = 3.74QLSAARR366 pKa = 11.84DD367 pKa = 3.84LIKK370 pKa = 10.51EE371 pKa = 3.93QWDD374 pKa = 3.98SVVGLDD380 pKa = 4.59IKK382 pKa = 10.73EE383 pKa = 3.96
Molecular weight: 40.72 kDa
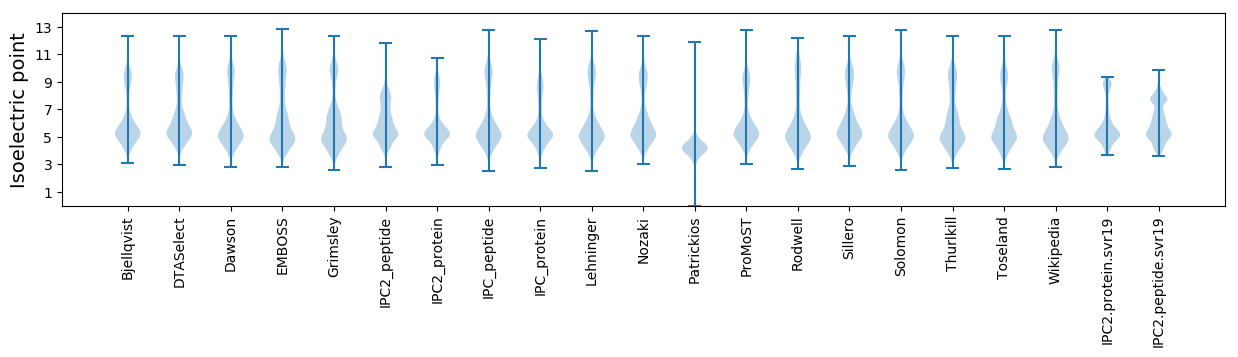
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A347ZNM1|A0A347ZNM1_9CHLR Exodeoxyribonuclease 7 small subunit OS=Pelolinea submarina OX=913107 GN=xseB PE=3 SV=1
MM1 pKa = 7.57KK2 pKa = 10.49NLARR6 pKa = 11.84PILICTVILLIAVLLGIGVGSISISPQEE34 pKa = 4.07SLSILWQALRR44 pKa = 11.84GLPLDD49 pKa = 4.69GEE51 pKa = 4.76GQSIPSVILLTLRR64 pKa = 11.84LPRR67 pKa = 11.84TLLMLLAGAALSGSGCAYY85 pKa = 10.13QGLFRR90 pKa = 11.84NPLADD95 pKa = 4.3PYY97 pKa = 11.14LIGAASGAGVGAVAAMSLNWPSTTLGYY124 pKa = 10.44LAVPLASFAGSLLAVFLVFRR144 pKa = 11.84LARR147 pKa = 11.84VGRR150 pKa = 11.84TLPVTNLILAGVAVSSFATALSSFMMVNAGGEE182 pKa = 4.02LRR184 pKa = 11.84RR185 pKa = 11.84AFVWMLGGSTMSGWKK200 pKa = 9.19PVLGMLPYY208 pKa = 10.18TLVGLGALLALSYY221 pKa = 10.91KK222 pKa = 10.83LNILQLGDD230 pKa = 3.61EE231 pKa = 4.5QAQQLGIKK239 pKa = 9.39VSRR242 pKa = 11.84VRR244 pKa = 11.84TQIILAATLATAAAVAFSGIIGFVGLIIPHH274 pKa = 5.11ITRR277 pKa = 11.84RR278 pKa = 11.84LWGGDD283 pKa = 3.49MRR285 pKa = 11.84RR286 pKa = 11.84MLPLSMLCGGAFLVFADD303 pKa = 3.44VLARR307 pKa = 11.84VIMAPQEE314 pKa = 4.1LPVGIITALCGAPFFLFIMRR334 pKa = 11.84QSKK337 pKa = 9.71EE338 pKa = 4.14GSWW341 pKa = 3.34
MM1 pKa = 7.57KK2 pKa = 10.49NLARR6 pKa = 11.84PILICTVILLIAVLLGIGVGSISISPQEE34 pKa = 4.07SLSILWQALRR44 pKa = 11.84GLPLDD49 pKa = 4.69GEE51 pKa = 4.76GQSIPSVILLTLRR64 pKa = 11.84LPRR67 pKa = 11.84TLLMLLAGAALSGSGCAYY85 pKa = 10.13QGLFRR90 pKa = 11.84NPLADD95 pKa = 4.3PYY97 pKa = 11.14LIGAASGAGVGAVAAMSLNWPSTTLGYY124 pKa = 10.44LAVPLASFAGSLLAVFLVFRR144 pKa = 11.84LARR147 pKa = 11.84VGRR150 pKa = 11.84TLPVTNLILAGVAVSSFATALSSFMMVNAGGEE182 pKa = 4.02LRR184 pKa = 11.84RR185 pKa = 11.84AFVWMLGGSTMSGWKK200 pKa = 9.19PVLGMLPYY208 pKa = 10.18TLVGLGALLALSYY221 pKa = 10.91KK222 pKa = 10.83LNILQLGDD230 pKa = 3.61EE231 pKa = 4.5QAQQLGIKK239 pKa = 9.39VSRR242 pKa = 11.84VRR244 pKa = 11.84TQIILAATLATAAAVAFSGIIGFVGLIIPHH274 pKa = 5.11ITRR277 pKa = 11.84RR278 pKa = 11.84LWGGDD283 pKa = 3.49MRR285 pKa = 11.84RR286 pKa = 11.84MLPLSMLCGGAFLVFADD303 pKa = 3.44VLARR307 pKa = 11.84VIMAPQEE314 pKa = 4.1LPVGIITALCGAPFFLFIMRR334 pKa = 11.84QSKK337 pKa = 9.71EE338 pKa = 4.14GSWW341 pKa = 3.34
Molecular weight: 35.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1049187 |
30 |
15355 |
342.2 |
37.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.721 ± 0.047 | 0.955 ± 0.021 |
5.624 ± 0.086 | 6.414 ± 0.058 |
4.207 ± 0.03 | 7.396 ± 0.101 |
1.816 ± 0.025 | 7.027 ± 0.039 |
4.778 ± 0.072 | 10.419 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.572 ± 0.037 | 4.112 ± 0.051 |
4.41 ± 0.056 | 3.897 ± 0.043 |
4.67 ± 0.073 | 6.183 ± 0.078 |
5.341 ± 0.087 | 6.86 ± 0.039 |
1.323 ± 0.022 | 3.276 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
